
Dragonfly associated cyclovirus 5
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Cyclovirus
Average proteome isoelectric point is 8.8
Get precalculated fractions of proteins
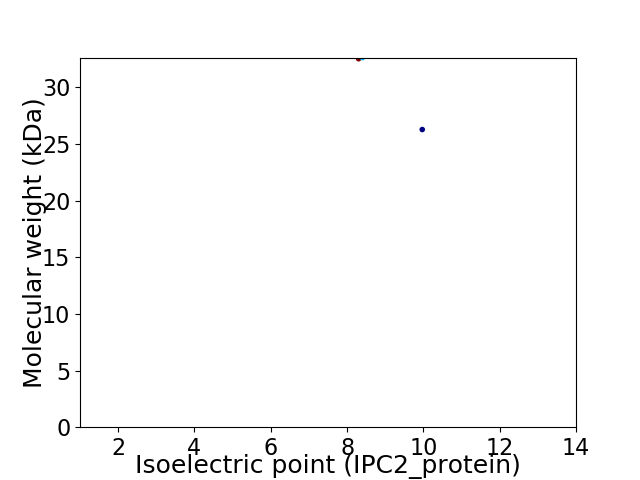
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K0A152|K0A152_9CIRC Putative capsid protein OS=Dragonfly associated cyclovirus 5 OX=1234883 PE=4 SV=1
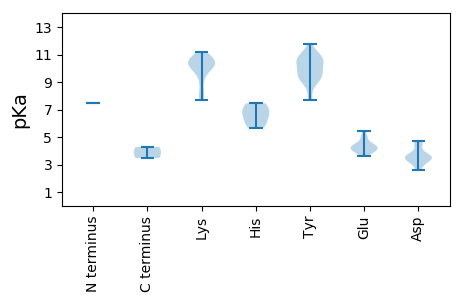
MM1 pKa = 7.49ANCVVRR7 pKa = 11.84RR8 pKa = 11.84FCFTLNNFTEE18 pKa = 4.23EE19 pKa = 4.34EE20 pKa = 4.46YY21 pKa = 11.11EE22 pKa = 4.05KK23 pKa = 10.17VTRR26 pKa = 11.84FIQDD30 pKa = 3.06YY31 pKa = 9.34CKK33 pKa = 10.81YY34 pKa = 10.13GIVGNEE40 pKa = 3.89TAPTTGTIHH49 pKa = 7.45LPGFCNLTKK58 pKa = 10.52GMRR61 pKa = 11.84FNNIKK66 pKa = 10.5SKK68 pKa = 10.37LATRR72 pKa = 11.84IHH74 pKa = 6.51LEE76 pKa = 3.96KK77 pKa = 11.17ANGTDD82 pKa = 3.41EE83 pKa = 4.22QNQIYY88 pKa = 9.43CRR90 pKa = 11.84KK91 pKa = 9.5SGTYY95 pKa = 9.25FEE97 pKa = 5.43KK98 pKa = 10.26GTPVGQGKK106 pKa = 7.85RR107 pKa = 11.84TDD109 pKa = 3.41LVSLVEE115 pKa = 5.0GIQNGQIRR123 pKa = 11.84LSDD126 pKa = 3.27IAKK129 pKa = 9.21DD130 pKa = 3.51HH131 pKa = 7.14PIAFIKK137 pKa = 7.71YY138 pKa = 7.66HH139 pKa = 6.07RR140 pKa = 11.84GIRR143 pKa = 11.84EE144 pKa = 3.8YY145 pKa = 11.1LQLTKK150 pKa = 10.21PVQPRR155 pKa = 11.84MFKK158 pKa = 8.93TWVYY162 pKa = 9.53YY163 pKa = 9.52YY164 pKa = 9.7WGPTGSGKK172 pKa = 9.98SSRR175 pKa = 11.84ALKK178 pKa = 10.0EE179 pKa = 3.61AMEE182 pKa = 4.5IEE184 pKa = 3.96GEE186 pKa = 4.07IYY188 pKa = 10.52YY189 pKa = 10.25KK190 pKa = 10.56PRR192 pKa = 11.84GLWWDD197 pKa = 4.71GYY199 pKa = 9.4HH200 pKa = 5.68QQDD203 pKa = 3.74NVIIDD208 pKa = 4.59DD209 pKa = 4.41FYY211 pKa = 11.73GWIKK215 pKa = 9.81YY216 pKa = 10.71DD217 pKa = 3.64EE218 pKa = 4.36MLKK221 pKa = 10.5IMDD224 pKa = 4.59RR225 pKa = 11.84YY226 pKa = 9.01PYY228 pKa = 9.88KK229 pKa = 10.71VQVKK233 pKa = 10.29GGFEE237 pKa = 4.2EE238 pKa = 4.82FTSRR242 pKa = 11.84RR243 pKa = 11.84IWITSNVDD251 pKa = 2.59TDD253 pKa = 3.55QLYY256 pKa = 10.61KK257 pKa = 10.85FIGYY261 pKa = 9.98VSDD264 pKa = 3.62AFDD267 pKa = 3.42RR268 pKa = 11.84RR269 pKa = 11.84ITNKK273 pKa = 9.82VYY275 pKa = 10.47IDD277 pKa = 3.47
MM1 pKa = 7.49ANCVVRR7 pKa = 11.84RR8 pKa = 11.84FCFTLNNFTEE18 pKa = 4.23EE19 pKa = 4.34EE20 pKa = 4.46YY21 pKa = 11.11EE22 pKa = 4.05KK23 pKa = 10.17VTRR26 pKa = 11.84FIQDD30 pKa = 3.06YY31 pKa = 9.34CKK33 pKa = 10.81YY34 pKa = 10.13GIVGNEE40 pKa = 3.89TAPTTGTIHH49 pKa = 7.45LPGFCNLTKK58 pKa = 10.52GMRR61 pKa = 11.84FNNIKK66 pKa = 10.5SKK68 pKa = 10.37LATRR72 pKa = 11.84IHH74 pKa = 6.51LEE76 pKa = 3.96KK77 pKa = 11.17ANGTDD82 pKa = 3.41EE83 pKa = 4.22QNQIYY88 pKa = 9.43CRR90 pKa = 11.84KK91 pKa = 9.5SGTYY95 pKa = 9.25FEE97 pKa = 5.43KK98 pKa = 10.26GTPVGQGKK106 pKa = 7.85RR107 pKa = 11.84TDD109 pKa = 3.41LVSLVEE115 pKa = 5.0GIQNGQIRR123 pKa = 11.84LSDD126 pKa = 3.27IAKK129 pKa = 9.21DD130 pKa = 3.51HH131 pKa = 7.14PIAFIKK137 pKa = 7.71YY138 pKa = 7.66HH139 pKa = 6.07RR140 pKa = 11.84GIRR143 pKa = 11.84EE144 pKa = 3.8YY145 pKa = 11.1LQLTKK150 pKa = 10.21PVQPRR155 pKa = 11.84MFKK158 pKa = 8.93TWVYY162 pKa = 9.53YY163 pKa = 9.52YY164 pKa = 9.7WGPTGSGKK172 pKa = 9.98SSRR175 pKa = 11.84ALKK178 pKa = 10.0EE179 pKa = 3.61AMEE182 pKa = 4.5IEE184 pKa = 3.96GEE186 pKa = 4.07IYY188 pKa = 10.52YY189 pKa = 10.25KK190 pKa = 10.56PRR192 pKa = 11.84GLWWDD197 pKa = 4.71GYY199 pKa = 9.4HH200 pKa = 5.68QQDD203 pKa = 3.74NVIIDD208 pKa = 4.59DD209 pKa = 4.41FYY211 pKa = 11.73GWIKK215 pKa = 9.81YY216 pKa = 10.71DD217 pKa = 3.64EE218 pKa = 4.36MLKK221 pKa = 10.5IMDD224 pKa = 4.59RR225 pKa = 11.84YY226 pKa = 9.01PYY228 pKa = 9.88KK229 pKa = 10.71VQVKK233 pKa = 10.29GGFEE237 pKa = 4.2EE238 pKa = 4.82FTSRR242 pKa = 11.84RR243 pKa = 11.84IWITSNVDD251 pKa = 2.59TDD253 pKa = 3.55QLYY256 pKa = 10.61KK257 pKa = 10.85FIGYY261 pKa = 9.98VSDD264 pKa = 3.62AFDD267 pKa = 3.42RR268 pKa = 11.84RR269 pKa = 11.84ITNKK273 pKa = 9.82VYY275 pKa = 10.47IDD277 pKa = 3.47
Molecular weight: 32.53 kDa
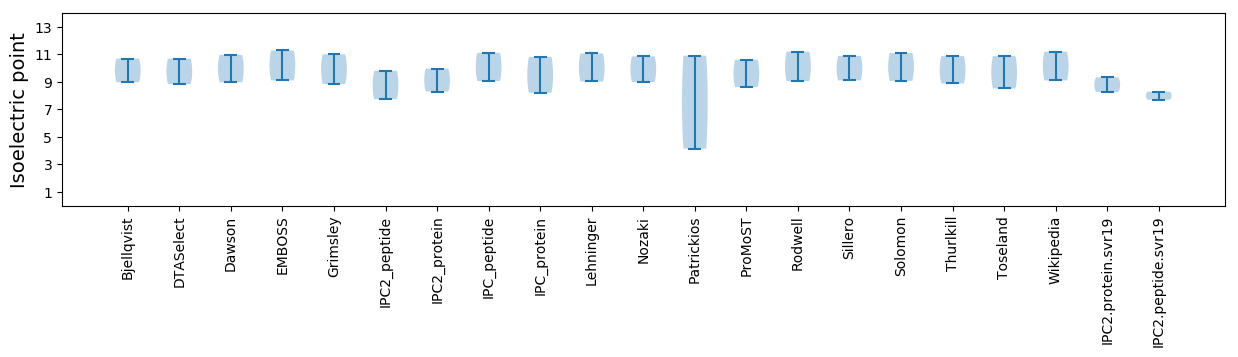
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K0A152|K0A152_9CIRC Putative capsid protein OS=Dragonfly associated cyclovirus 5 OX=1234883 PE=4 SV=1
MM1 pKa = 7.48ARR3 pKa = 11.84FKK5 pKa = 10.86RR6 pKa = 11.84VFRR9 pKa = 11.84RR10 pKa = 11.84PRR12 pKa = 11.84RR13 pKa = 11.84VVRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84SRR20 pKa = 11.84MFRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84YY26 pKa = 8.9RR27 pKa = 11.84RR28 pKa = 11.84TRR30 pKa = 11.84KK31 pKa = 8.47RR32 pKa = 11.84TSGTMWCKK40 pKa = 8.19LTKK43 pKa = 10.37VATVQVKK50 pKa = 10.38NDD52 pKa = 3.56STSVWSGSFYY62 pKa = 10.7PNDD65 pKa = 3.24FPEE68 pKa = 4.52YY69 pKa = 10.63VSLAPNFEE77 pKa = 5.07AIQFVKK83 pKa = 9.95MRR85 pKa = 11.84MRR87 pKa = 11.84VMPLQNVSNNSTSQTPAYY105 pKa = 10.68AMLPWHH111 pKa = 6.81KK112 pKa = 10.58GGPLQKK118 pKa = 10.72AFNTYY123 pKa = 10.81LSVDD127 pKa = 2.62KK128 pKa = 11.07CKK130 pKa = 10.2IYY132 pKa = 10.72RR133 pKa = 11.84QTQQGYY139 pKa = 7.67QSYY142 pKa = 9.7KK143 pKa = 10.64CSVLQNAFTKK153 pKa = 10.34SSSAISEE160 pKa = 4.34DD161 pKa = 3.58FSLATVTKK169 pKa = 7.79WCPRR173 pKa = 11.84IEE175 pKa = 4.35RR176 pKa = 11.84PNNMEE181 pKa = 3.96AKK183 pKa = 9.88QPQIYY188 pKa = 8.89TGIVVFQGEE197 pKa = 4.14NTMAGRR203 pKa = 11.84TTAFNIIQDD212 pKa = 3.59VWVKK216 pKa = 10.46CINQSNFSII225 pKa = 4.27
MM1 pKa = 7.48ARR3 pKa = 11.84FKK5 pKa = 10.86RR6 pKa = 11.84VFRR9 pKa = 11.84RR10 pKa = 11.84PRR12 pKa = 11.84RR13 pKa = 11.84VVRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84SRR20 pKa = 11.84MFRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84YY26 pKa = 8.9RR27 pKa = 11.84RR28 pKa = 11.84TRR30 pKa = 11.84KK31 pKa = 8.47RR32 pKa = 11.84TSGTMWCKK40 pKa = 8.19LTKK43 pKa = 10.37VATVQVKK50 pKa = 10.38NDD52 pKa = 3.56STSVWSGSFYY62 pKa = 10.7PNDD65 pKa = 3.24FPEE68 pKa = 4.52YY69 pKa = 10.63VSLAPNFEE77 pKa = 5.07AIQFVKK83 pKa = 9.95MRR85 pKa = 11.84MRR87 pKa = 11.84VMPLQNVSNNSTSQTPAYY105 pKa = 10.68AMLPWHH111 pKa = 6.81KK112 pKa = 10.58GGPLQKK118 pKa = 10.72AFNTYY123 pKa = 10.81LSVDD127 pKa = 2.62KK128 pKa = 11.07CKK130 pKa = 10.2IYY132 pKa = 10.72RR133 pKa = 11.84QTQQGYY139 pKa = 7.67QSYY142 pKa = 9.7KK143 pKa = 10.64CSVLQNAFTKK153 pKa = 10.34SSSAISEE160 pKa = 4.34DD161 pKa = 3.58FSLATVTKK169 pKa = 7.79WCPRR173 pKa = 11.84IEE175 pKa = 4.35RR176 pKa = 11.84PNNMEE181 pKa = 3.96AKK183 pKa = 9.88QPQIYY188 pKa = 8.89TGIVVFQGEE197 pKa = 4.14NTMAGRR203 pKa = 11.84TTAFNIIQDD212 pKa = 3.59VWVKK216 pKa = 10.46CINQSNFSII225 pKa = 4.27
Molecular weight: 26.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
502 |
225 |
277 |
251.0 |
29.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.382 ± 0.884 | 1.992 ± 0.146 |
4.183 ± 1.242 | 4.582 ± 1.213 |
5.378 ± 0.253 | 6.175 ± 1.659 |
1.195 ± 0.476 | 6.574 ± 1.349 |
7.371 ± 0.446 | 4.382 ± 0.524 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.988 ± 0.641 | 5.378 ± 0.534 |
3.984 ± 0.573 | 5.378 ± 0.816 |
8.167 ± 1.302 | 5.976 ± 1.845 |
7.371 ± 0.117 | 6.574 ± 0.903 |
2.191 ± 0.02 | 5.777 ± 1.125 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
