
Plesiocystis pacifica SIR-1
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Myxococcales; Nannocystineae; Nannocystaceae; Plesiocystis; Plesiocystis pacifica
Average proteome isoelectric point is 5.8
Get precalculated fractions of proteins
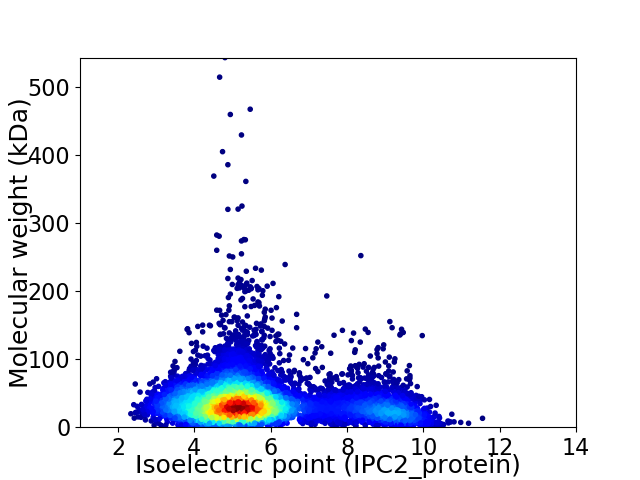
Virtual 2D-PAGE plot for 8437 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A6GBS3|A6GBS3_9DELT Uncharacterized protein OS=Plesiocystis pacifica SIR-1 OX=391625 GN=PPSIR1_38219 PE=4 SV=1
MM1 pKa = 7.52SSPARR6 pKa = 11.84SLALTFTASLACLALSLFACADD28 pKa = 4.05DD29 pKa = 5.53DD30 pKa = 5.02PPLGDD35 pKa = 4.87PPPNSPGGGGTLTAGGGDD53 pKa = 3.46GGGGGTLTAGGGGDD67 pKa = 4.18DD68 pKa = 4.93EE69 pKa = 5.15VGEE72 pKa = 5.17DD73 pKa = 5.0DD74 pKa = 4.1PCSDD78 pKa = 4.44LGSTAVDD85 pKa = 5.48FSYY88 pKa = 10.85LWVANTDD95 pKa = 3.38EE96 pKa = 4.95GSVSKK101 pKa = 11.16VNAKK105 pKa = 8.03TQVEE109 pKa = 4.44VARR112 pKa = 11.84YY113 pKa = 6.51RR114 pKa = 11.84TGPANVEE121 pKa = 4.1HH122 pKa = 6.65SPSRR126 pKa = 11.84TTVSLDD132 pKa = 2.8GRR134 pKa = 11.84YY135 pKa = 10.08ALVGNRR141 pKa = 11.84LSGSVVLVAAEE152 pKa = 4.15TGDD155 pKa = 3.98CVDD158 pKa = 3.98ANEE161 pKa = 5.37DD162 pKa = 3.74GLITTSQSPDD172 pKa = 3.78DD173 pKa = 4.24LLDD176 pKa = 3.67WGDD179 pKa = 4.19DD180 pKa = 3.7EE181 pKa = 5.54CVLWSTNMPQIGEE194 pKa = 4.52GISAGPRR201 pKa = 11.84GMTFDD206 pKa = 4.36PGEE209 pKa = 5.36LDD211 pKa = 3.64TQTCTYY217 pKa = 10.09QDD219 pKa = 3.06AKK221 pKa = 10.73IWVGWGGASTDD232 pKa = 3.53LAHH235 pKa = 7.78IGRR238 pKa = 11.84LDD240 pKa = 3.67PATGTFEE247 pKa = 4.14EE248 pKa = 5.38LIAVEE253 pKa = 4.73DD254 pKa = 3.94WGSSLGPVHH263 pKa = 6.94APYY266 pKa = 10.54GAAHH270 pKa = 7.13DD271 pKa = 4.8GDD273 pKa = 3.82HH274 pKa = 6.5SVWFTGLRR282 pKa = 11.84GEE284 pKa = 4.4YY285 pKa = 10.46LRR287 pKa = 11.84IDD289 pKa = 3.93TEE291 pKa = 4.3DD292 pKa = 4.57LDD294 pKa = 3.88VDD296 pKa = 3.75RR297 pKa = 11.84WIAPGWAAPYY307 pKa = 10.74GMAVDD312 pKa = 3.6AEE314 pKa = 4.36GRR316 pKa = 11.84VWTGGCFGPVSVFDD330 pKa = 4.83PDD332 pKa = 3.93TEE334 pKa = 4.23IVTEE338 pKa = 4.09IVGTDD343 pKa = 2.83ACHH346 pKa = 6.72RR347 pKa = 11.84GVAADD352 pKa = 3.46TKK354 pKa = 9.81GNVWVAMNEE363 pKa = 3.96PCGLVQIDD371 pKa = 3.81RR372 pKa = 11.84DD373 pKa = 3.86DD374 pKa = 4.28EE375 pKa = 4.39EE376 pKa = 4.74VVNVLTAEE384 pKa = 4.58DD385 pKa = 4.44FDD387 pKa = 4.71GLCSIPVGVSVDD399 pKa = 3.23ADD401 pKa = 3.69DD402 pKa = 4.3FVWMVDD408 pKa = 3.3QLGWAWRR415 pKa = 11.84IDD417 pKa = 3.96PEE419 pKa = 4.51SLDD422 pKa = 3.48MDD424 pKa = 4.17YY425 pKa = 11.79LEE427 pKa = 5.42IPGNHH432 pKa = 5.1YY433 pKa = 9.77TYY435 pKa = 11.62SDD437 pKa = 3.46MTGGGLNAVVYY448 pKa = 8.07PQQ450 pKa = 3.24
MM1 pKa = 7.52SSPARR6 pKa = 11.84SLALTFTASLACLALSLFACADD28 pKa = 4.05DD29 pKa = 5.53DD30 pKa = 5.02PPLGDD35 pKa = 4.87PPPNSPGGGGTLTAGGGDD53 pKa = 3.46GGGGGTLTAGGGGDD67 pKa = 4.18DD68 pKa = 4.93EE69 pKa = 5.15VGEE72 pKa = 5.17DD73 pKa = 5.0DD74 pKa = 4.1PCSDD78 pKa = 4.44LGSTAVDD85 pKa = 5.48FSYY88 pKa = 10.85LWVANTDD95 pKa = 3.38EE96 pKa = 4.95GSVSKK101 pKa = 11.16VNAKK105 pKa = 8.03TQVEE109 pKa = 4.44VARR112 pKa = 11.84YY113 pKa = 6.51RR114 pKa = 11.84TGPANVEE121 pKa = 4.1HH122 pKa = 6.65SPSRR126 pKa = 11.84TTVSLDD132 pKa = 2.8GRR134 pKa = 11.84YY135 pKa = 10.08ALVGNRR141 pKa = 11.84LSGSVVLVAAEE152 pKa = 4.15TGDD155 pKa = 3.98CVDD158 pKa = 3.98ANEE161 pKa = 5.37DD162 pKa = 3.74GLITTSQSPDD172 pKa = 3.78DD173 pKa = 4.24LLDD176 pKa = 3.67WGDD179 pKa = 4.19DD180 pKa = 3.7EE181 pKa = 5.54CVLWSTNMPQIGEE194 pKa = 4.52GISAGPRR201 pKa = 11.84GMTFDD206 pKa = 4.36PGEE209 pKa = 5.36LDD211 pKa = 3.64TQTCTYY217 pKa = 10.09QDD219 pKa = 3.06AKK221 pKa = 10.73IWVGWGGASTDD232 pKa = 3.53LAHH235 pKa = 7.78IGRR238 pKa = 11.84LDD240 pKa = 3.67PATGTFEE247 pKa = 4.14EE248 pKa = 5.38LIAVEE253 pKa = 4.73DD254 pKa = 3.94WGSSLGPVHH263 pKa = 6.94APYY266 pKa = 10.54GAAHH270 pKa = 7.13DD271 pKa = 4.8GDD273 pKa = 3.82HH274 pKa = 6.5SVWFTGLRR282 pKa = 11.84GEE284 pKa = 4.4YY285 pKa = 10.46LRR287 pKa = 11.84IDD289 pKa = 3.93TEE291 pKa = 4.3DD292 pKa = 4.57LDD294 pKa = 3.88VDD296 pKa = 3.75RR297 pKa = 11.84WIAPGWAAPYY307 pKa = 10.74GMAVDD312 pKa = 3.6AEE314 pKa = 4.36GRR316 pKa = 11.84VWTGGCFGPVSVFDD330 pKa = 4.83PDD332 pKa = 3.93TEE334 pKa = 4.23IVTEE338 pKa = 4.09IVGTDD343 pKa = 2.83ACHH346 pKa = 6.72RR347 pKa = 11.84GVAADD352 pKa = 3.46TKK354 pKa = 9.81GNVWVAMNEE363 pKa = 3.96PCGLVQIDD371 pKa = 3.81RR372 pKa = 11.84DD373 pKa = 3.86DD374 pKa = 4.28EE375 pKa = 4.39EE376 pKa = 4.74VVNVLTAEE384 pKa = 4.58DD385 pKa = 4.44FDD387 pKa = 4.71GLCSIPVGVSVDD399 pKa = 3.23ADD401 pKa = 3.69DD402 pKa = 4.3FVWMVDD408 pKa = 3.3QLGWAWRR415 pKa = 11.84IDD417 pKa = 3.96PEE419 pKa = 4.51SLDD422 pKa = 3.48MDD424 pKa = 4.17YY425 pKa = 11.79LEE427 pKa = 5.42IPGNHH432 pKa = 5.1YY433 pKa = 9.77TYY435 pKa = 11.62SDD437 pKa = 3.46MTGGGLNAVVYY448 pKa = 8.07PQQ450 pKa = 3.24
Molecular weight: 47.2 kDa
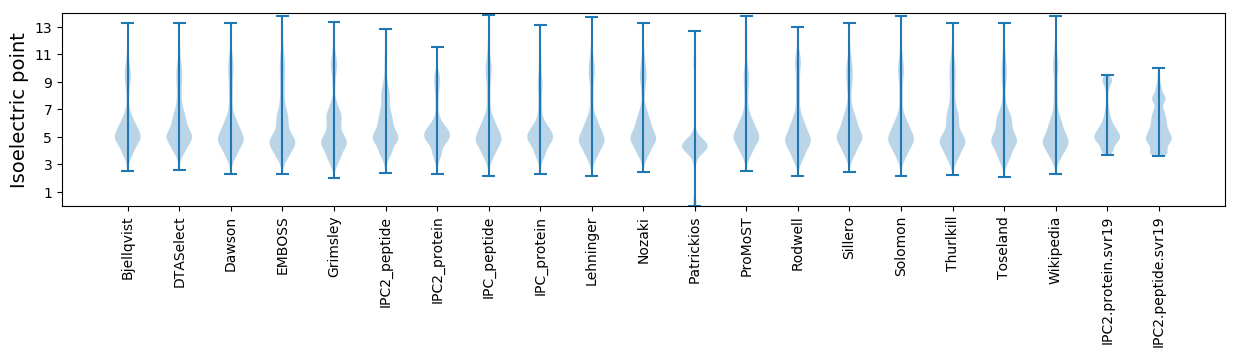
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A6GEB9|A6GEB9_9DELT (2Fe-2S)-binding protein OS=Plesiocystis pacifica SIR-1 OX=391625 GN=PPSIR1_17675 PE=4 SV=1
MM1 pKa = 7.74RR2 pKa = 11.84RR3 pKa = 11.84RR4 pKa = 11.84APSTASRR11 pKa = 11.84RR12 pKa = 11.84AGVHH16 pKa = 5.25SSAMAAPAPAQGSTGAAGSAPPTRR40 pKa = 11.84AKK42 pKa = 10.5LSSAPAPAVPTPMAASHH59 pKa = 5.88RR60 pKa = 11.84RR61 pKa = 11.84LRR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84ARR67 pKa = 11.84GSARR71 pKa = 11.84RR72 pKa = 11.84AASSSAMVGKK82 pKa = 10.47RR83 pKa = 11.84SSTSTSRR90 pKa = 11.84ARR92 pKa = 11.84SRR94 pKa = 11.84ATSTWLGTFASRR106 pKa = 11.84FVSLLASLGGARR118 pKa = 11.84RR119 pKa = 11.84WPSRR123 pKa = 11.84TAMAA127 pKa = 5.11
MM1 pKa = 7.74RR2 pKa = 11.84RR3 pKa = 11.84RR4 pKa = 11.84APSTASRR11 pKa = 11.84RR12 pKa = 11.84AGVHH16 pKa = 5.25SSAMAAPAPAQGSTGAAGSAPPTRR40 pKa = 11.84AKK42 pKa = 10.5LSSAPAPAVPTPMAASHH59 pKa = 5.88RR60 pKa = 11.84RR61 pKa = 11.84LRR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84ARR67 pKa = 11.84GSARR71 pKa = 11.84RR72 pKa = 11.84AASSSAMVGKK82 pKa = 10.47RR83 pKa = 11.84SSTSTSRR90 pKa = 11.84ARR92 pKa = 11.84SRR94 pKa = 11.84ATSTWLGTFASRR106 pKa = 11.84FVSLLASLGGARR118 pKa = 11.84RR119 pKa = 11.84WPSRR123 pKa = 11.84TAMAA127 pKa = 5.11
Molecular weight: 13.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3147198 |
17 |
4933 |
373.0 |
40.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.675 ± 0.037 | 1.225 ± 0.015 |
6.439 ± 0.024 | 7.553 ± 0.026 |
3.082 ± 0.016 | 9.064 ± 0.029 |
2.109 ± 0.013 | 3.391 ± 0.019 |
2.303 ± 0.023 | 10.505 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.845 ± 0.011 | 1.941 ± 0.013 |
5.868 ± 0.021 | 2.918 ± 0.013 |
7.698 ± 0.036 | 5.594 ± 0.02 |
4.952 ± 0.023 | 7.32 ± 0.02 |
1.537 ± 0.01 | 1.983 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
