
Monkey B-lymphotropic papovavirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; unclassified Polyomaviridae; African green monkey polyomavirus
Average proteome isoelectric point is 7.02
Get precalculated fractions of proteins
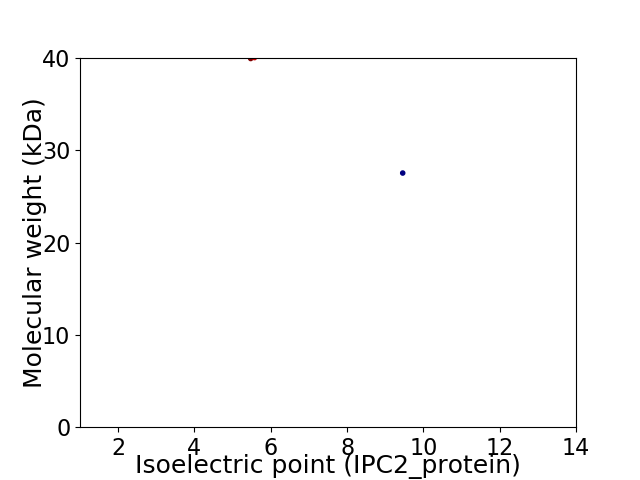
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
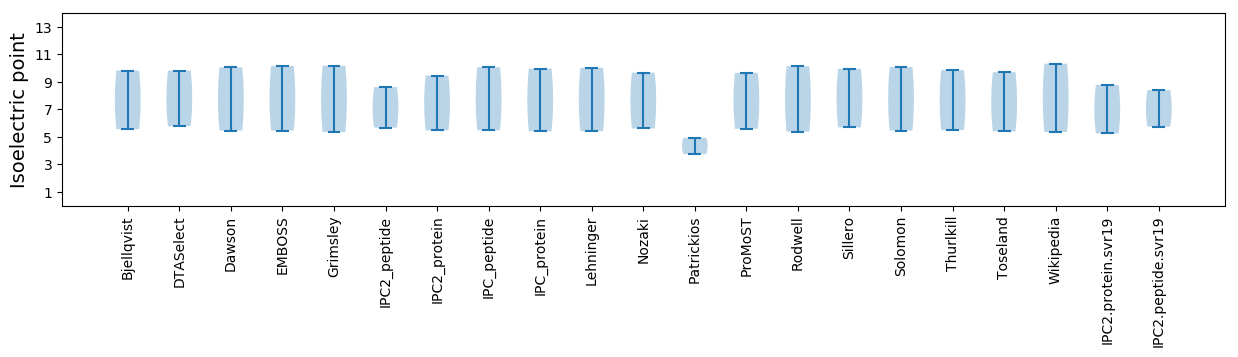
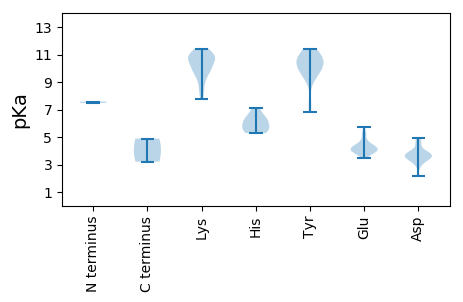
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q90131|Q90131_9POLY Minor capsid protein OS=Monkey B-lymphotropic papovavirus OX=10574 PE=3 SV=1
MM1 pKa = 7.58APQRR5 pKa = 11.84KK6 pKa = 8.36RR7 pKa = 11.84QDD9 pKa = 3.48GACKK13 pKa = 8.94KK14 pKa = 7.79TCPIPAPVPRR24 pKa = 11.84LLVKK28 pKa = 10.8GGVEE32 pKa = 3.97VLEE35 pKa = 4.41VRR37 pKa = 11.84TGPDD41 pKa = 3.47AITQIEE47 pKa = 4.77AYY49 pKa = 10.29LNPRR53 pKa = 11.84MGNNIPSEE61 pKa = 4.0DD62 pKa = 3.61LYY64 pKa = 11.38GYY66 pKa = 10.56SNSINTAFSKK76 pKa = 11.09ASDD79 pKa = 3.87TPNKK83 pKa = 8.68GTLPCYY89 pKa = 10.03SVAVIKK95 pKa = 10.87LPLLNEE101 pKa = 4.73DD102 pKa = 3.9MTCDD106 pKa = 3.74TILMWEE112 pKa = 4.1AVSVKK117 pKa = 9.88TEE119 pKa = 3.84VVGISSLVNLHH130 pKa = 6.16QGGKK134 pKa = 9.56YY135 pKa = 9.87IYY137 pKa = 9.33GSSSGCVPVQGTTYY151 pKa = 11.11HH152 pKa = 5.69MFAVGGEE159 pKa = 3.95PLEE162 pKa = 4.2LQGLVASSTATYY174 pKa = 9.73PDD176 pKa = 3.86DD177 pKa = 3.58VVAIKK182 pKa = 10.66NMKK185 pKa = 9.52PGNQGLDD192 pKa = 3.51PKK194 pKa = 11.16AKK196 pKa = 10.51ALLDD200 pKa = 3.67KK201 pKa = 10.9DD202 pKa = 3.32GKK204 pKa = 10.91YY205 pKa = 9.68PVEE208 pKa = 4.34VWCPDD213 pKa = 3.3PSKK216 pKa = 11.43NEE218 pKa = 3.45NTRR221 pKa = 11.84YY222 pKa = 10.01YY223 pKa = 11.35GSFTGGATTPPVMQFTNSVTTVLLDD248 pKa = 3.75EE249 pKa = 5.12NGVGPLCKK257 pKa = 9.66GDD259 pKa = 4.92KK260 pKa = 10.77LFLSCADD267 pKa = 3.05IAGVHH272 pKa = 5.36TNYY275 pKa = 10.39SEE277 pKa = 4.22TQSWRR282 pKa = 11.84GLPRR286 pKa = 11.84YY287 pKa = 9.97FNVTLRR293 pKa = 11.84KK294 pKa = 9.82RR295 pKa = 11.84IVKK298 pKa = 9.99NPYY301 pKa = 9.02PVSSLLNSFFSGLMPQIQGQPMEE324 pKa = 4.52GVSGQVEE331 pKa = 4.1EE332 pKa = 4.1VRR334 pKa = 11.84IFEE337 pKa = 4.29GTEE340 pKa = 3.96GLPGDD345 pKa = 4.53PDD347 pKa = 3.8LNRR350 pKa = 11.84YY351 pKa = 7.58VDD353 pKa = 4.7KK354 pKa = 10.92FCQNQTVLPVSNDD367 pKa = 2.95MM368 pKa = 4.89
MM1 pKa = 7.58APQRR5 pKa = 11.84KK6 pKa = 8.36RR7 pKa = 11.84QDD9 pKa = 3.48GACKK13 pKa = 8.94KK14 pKa = 7.79TCPIPAPVPRR24 pKa = 11.84LLVKK28 pKa = 10.8GGVEE32 pKa = 3.97VLEE35 pKa = 4.41VRR37 pKa = 11.84TGPDD41 pKa = 3.47AITQIEE47 pKa = 4.77AYY49 pKa = 10.29LNPRR53 pKa = 11.84MGNNIPSEE61 pKa = 4.0DD62 pKa = 3.61LYY64 pKa = 11.38GYY66 pKa = 10.56SNSINTAFSKK76 pKa = 11.09ASDD79 pKa = 3.87TPNKK83 pKa = 8.68GTLPCYY89 pKa = 10.03SVAVIKK95 pKa = 10.87LPLLNEE101 pKa = 4.73DD102 pKa = 3.9MTCDD106 pKa = 3.74TILMWEE112 pKa = 4.1AVSVKK117 pKa = 9.88TEE119 pKa = 3.84VVGISSLVNLHH130 pKa = 6.16QGGKK134 pKa = 9.56YY135 pKa = 9.87IYY137 pKa = 9.33GSSSGCVPVQGTTYY151 pKa = 11.11HH152 pKa = 5.69MFAVGGEE159 pKa = 3.95PLEE162 pKa = 4.2LQGLVASSTATYY174 pKa = 9.73PDD176 pKa = 3.86DD177 pKa = 3.58VVAIKK182 pKa = 10.66NMKK185 pKa = 9.52PGNQGLDD192 pKa = 3.51PKK194 pKa = 11.16AKK196 pKa = 10.51ALLDD200 pKa = 3.67KK201 pKa = 10.9DD202 pKa = 3.32GKK204 pKa = 10.91YY205 pKa = 9.68PVEE208 pKa = 4.34VWCPDD213 pKa = 3.3PSKK216 pKa = 11.43NEE218 pKa = 3.45NTRR221 pKa = 11.84YY222 pKa = 10.01YY223 pKa = 11.35GSFTGGATTPPVMQFTNSVTTVLLDD248 pKa = 3.75EE249 pKa = 5.12NGVGPLCKK257 pKa = 9.66GDD259 pKa = 4.92KK260 pKa = 10.77LFLSCADD267 pKa = 3.05IAGVHH272 pKa = 5.36TNYY275 pKa = 10.39SEE277 pKa = 4.22TQSWRR282 pKa = 11.84GLPRR286 pKa = 11.84YY287 pKa = 9.97FNVTLRR293 pKa = 11.84KK294 pKa = 9.82RR295 pKa = 11.84IVKK298 pKa = 9.99NPYY301 pKa = 9.02PVSSLLNSFFSGLMPQIQGQPMEE324 pKa = 4.52GVSGQVEE331 pKa = 4.1EE332 pKa = 4.1VRR334 pKa = 11.84IFEE337 pKa = 4.29GTEE340 pKa = 3.96GLPGDD345 pKa = 4.53PDD347 pKa = 3.8LNRR350 pKa = 11.84YY351 pKa = 7.58VDD353 pKa = 4.7KK354 pKa = 10.92FCQNQTVLPVSNDD367 pKa = 2.95MM368 pKa = 4.89
Molecular weight: 39.93 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q90131|Q90131_9POLY Minor capsid protein OS=Monkey B-lymphotropic papovavirus OX=10574 PE=3 SV=1
MM1 pKa = 7.48ALVPWFPQVDD11 pKa = 3.83YY12 pKa = 10.54LFPGFTSFSYY22 pKa = 10.85YY23 pKa = 10.78LNAVLDD29 pKa = 3.69WGEE32 pKa = 4.36SLFHH36 pKa = 6.37AVGRR40 pKa = 11.84EE41 pKa = 4.04VWRR44 pKa = 11.84HH45 pKa = 5.29LMRR48 pKa = 11.84QATLQIGQATRR59 pKa = 11.84AVAVRR64 pKa = 11.84STNEE68 pKa = 3.61LSHH71 pKa = 6.47TLAQIAEE78 pKa = 4.22NARR81 pKa = 11.84WALTSGPVHH90 pKa = 7.08IYY92 pKa = 10.89SSVQDD97 pKa = 3.52YY98 pKa = 10.9YY99 pKa = 11.36RR100 pKa = 11.84YY101 pKa = 10.31LPARR105 pKa = 11.84NPIQLRR111 pKa = 11.84QEE113 pKa = 3.8YY114 pKa = 9.42RR115 pKa = 11.84NRR117 pKa = 11.84GEE119 pKa = 4.36PPPSRR124 pKa = 11.84ADD126 pKa = 3.39FEE128 pKa = 4.47YY129 pKa = 10.79QEE131 pKa = 4.08NRR133 pKa = 11.84EE134 pKa = 4.12GQRR137 pKa = 11.84ARR139 pKa = 11.84RR140 pKa = 11.84EE141 pKa = 3.93LGYY144 pKa = 10.58DD145 pKa = 3.48EE146 pKa = 5.27PRR148 pKa = 11.84SGQYY152 pKa = 9.54VEE154 pKa = 5.75HH155 pKa = 5.94YY156 pKa = 6.84TAPGGAHH163 pKa = 5.58QRR165 pKa = 11.84VTQDD169 pKa = 2.16WMLPLILGLYY179 pKa = 10.53GDD181 pKa = 4.55ITPTWEE187 pKa = 3.77VEE189 pKa = 4.03LNKK192 pKa = 10.48LEE194 pKa = 4.31KK195 pKa = 11.11EE196 pKa = 3.78EE197 pKa = 5.3DD198 pKa = 3.85GPSKK202 pKa = 10.61KK203 pKa = 9.93KK204 pKa = 10.56ARR206 pKa = 11.84RR207 pKa = 11.84SMQKK211 pKa = 9.95NMPYY215 pKa = 10.19SRR217 pKa = 11.84SRR219 pKa = 11.84PQAPSKK225 pKa = 10.27RR226 pKa = 11.84RR227 pKa = 11.84SRR229 pKa = 11.84GARR232 pKa = 11.84SKK234 pKa = 11.07NRR236 pKa = 11.84AA237 pKa = 3.21
MM1 pKa = 7.48ALVPWFPQVDD11 pKa = 3.83YY12 pKa = 10.54LFPGFTSFSYY22 pKa = 10.85YY23 pKa = 10.78LNAVLDD29 pKa = 3.69WGEE32 pKa = 4.36SLFHH36 pKa = 6.37AVGRR40 pKa = 11.84EE41 pKa = 4.04VWRR44 pKa = 11.84HH45 pKa = 5.29LMRR48 pKa = 11.84QATLQIGQATRR59 pKa = 11.84AVAVRR64 pKa = 11.84STNEE68 pKa = 3.61LSHH71 pKa = 6.47TLAQIAEE78 pKa = 4.22NARR81 pKa = 11.84WALTSGPVHH90 pKa = 7.08IYY92 pKa = 10.89SSVQDD97 pKa = 3.52YY98 pKa = 10.9YY99 pKa = 11.36RR100 pKa = 11.84YY101 pKa = 10.31LPARR105 pKa = 11.84NPIQLRR111 pKa = 11.84QEE113 pKa = 3.8YY114 pKa = 9.42RR115 pKa = 11.84NRR117 pKa = 11.84GEE119 pKa = 4.36PPPSRR124 pKa = 11.84ADD126 pKa = 3.39FEE128 pKa = 4.47YY129 pKa = 10.79QEE131 pKa = 4.08NRR133 pKa = 11.84EE134 pKa = 4.12GQRR137 pKa = 11.84ARR139 pKa = 11.84RR140 pKa = 11.84EE141 pKa = 3.93LGYY144 pKa = 10.58DD145 pKa = 3.48EE146 pKa = 5.27PRR148 pKa = 11.84SGQYY152 pKa = 9.54VEE154 pKa = 5.75HH155 pKa = 5.94YY156 pKa = 6.84TAPGGAHH163 pKa = 5.58QRR165 pKa = 11.84VTQDD169 pKa = 2.16WMLPLILGLYY179 pKa = 10.53GDD181 pKa = 4.55ITPTWEE187 pKa = 3.77VEE189 pKa = 4.03LNKK192 pKa = 10.48LEE194 pKa = 4.31KK195 pKa = 11.11EE196 pKa = 3.78EE197 pKa = 5.3DD198 pKa = 3.85GPSKK202 pKa = 10.61KK203 pKa = 9.93KK204 pKa = 10.56ARR206 pKa = 11.84RR207 pKa = 11.84SMQKK211 pKa = 9.95NMPYY215 pKa = 10.19SRR217 pKa = 11.84SRR219 pKa = 11.84PQAPSKK225 pKa = 10.27RR226 pKa = 11.84RR227 pKa = 11.84SRR229 pKa = 11.84GARR232 pKa = 11.84SKK234 pKa = 11.07NRR236 pKa = 11.84AA237 pKa = 3.21
Molecular weight: 27.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
605 |
237 |
368 |
302.5 |
33.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.281 ± 1.17 | 1.488 ± 0.806 |
4.463 ± 0.589 | 5.785 ± 0.752 |
2.645 ± 0.061 | 8.099 ± 0.96 |
1.488 ± 0.566 | 3.306 ± 0.42 |
4.793 ± 0.769 | 8.264 ± 0.134 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.479 ± 0.2 | 4.959 ± 0.63 |
7.769 ± 0.323 | 4.959 ± 0.743 |
6.446 ± 2.681 | 7.107 ± 0.036 |
5.785 ± 0.849 | 7.603 ± 1.377 |
1.488 ± 0.566 | 4.793 ± 0.604 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
