
Beihai weivirus-like virus 9
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.34
Get precalculated fractions of proteins
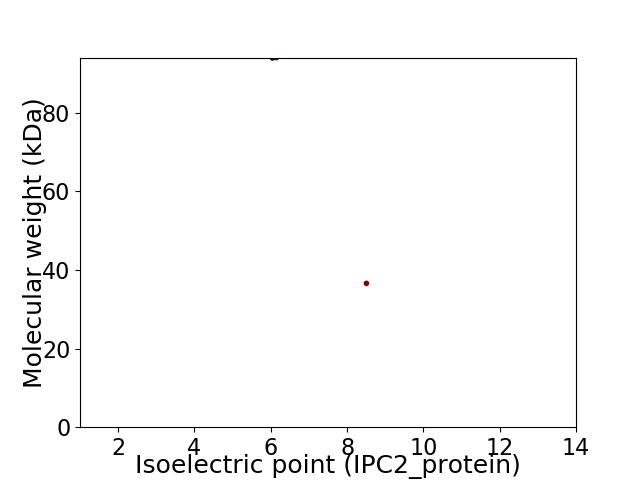
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KLA9|A0A1L3KLA9_9VIRU RNA-dependent RNA polymerase OS=Beihai weivirus-like virus 9 OX=1922756 PE=4 SV=1
MM1 pKa = 7.49TLPLSVANTTSLRR14 pKa = 11.84DD15 pKa = 3.27IEE17 pKa = 4.42GWMFKK22 pKa = 10.04IASEE26 pKa = 4.26RR27 pKa = 11.84PGMYY31 pKa = 9.49TIRR34 pKa = 11.84VRR36 pKa = 11.84LPQRR40 pKa = 11.84RR41 pKa = 11.84CGSCIKK47 pKa = 9.75LTRR50 pKa = 11.84SPLDD54 pKa = 3.56DD55 pKa = 3.54MEE57 pKa = 4.35EE58 pKa = 4.36VVVHH62 pKa = 5.08ITGEE66 pKa = 4.13EE67 pKa = 3.86FAKK70 pKa = 10.25LDD72 pKa = 3.83KK73 pKa = 11.27VIMNQSYY80 pKa = 10.49SEE82 pKa = 4.38SKK84 pKa = 10.48PKK86 pKa = 10.1VQAAIIYY93 pKa = 7.7QHH95 pKa = 7.13ASTIWPSHH103 pKa = 5.74TPDD106 pKa = 3.84DD107 pKa = 3.72VTMRR111 pKa = 11.84TLGAWAITQYY121 pKa = 10.67RR122 pKa = 11.84CNGGWAKK129 pKa = 10.66RR130 pKa = 11.84LLKK133 pKa = 10.3CIPCLQYY140 pKa = 9.57MQKK143 pKa = 10.16EE144 pKa = 4.11VHH146 pKa = 6.71LDD148 pKa = 3.57LKK150 pKa = 9.91GLRR153 pKa = 11.84MPNRR157 pKa = 11.84PVPPTPVEE165 pKa = 4.24SEE167 pKa = 4.01EE168 pKa = 4.26SSAGGTNGSTVRR180 pKa = 11.84TNTTATTAEE189 pKa = 4.82LSSQPTVEE197 pKa = 4.25SLPTITEE204 pKa = 3.97HH205 pKa = 8.08SDD207 pKa = 3.46EE208 pKa = 4.23EE209 pKa = 4.7DD210 pKa = 3.17EE211 pKa = 4.22NARR214 pKa = 11.84ANEE217 pKa = 4.28TPGEE221 pKa = 3.88AWARR225 pKa = 11.84WLAAGPPRR233 pKa = 11.84NEE235 pKa = 3.32IGYY238 pKa = 9.76VEE240 pKa = 4.87LGHH243 pKa = 6.84GNEE246 pKa = 4.75DD247 pKa = 3.61LHH249 pKa = 7.35PLGGASAEE257 pKa = 4.16DD258 pKa = 3.36SAIRR262 pKa = 11.84AEE264 pKa = 4.18RR265 pKa = 11.84QDD267 pKa = 3.48RR268 pKa = 11.84VLEE271 pKa = 4.04NAAGVGVVGQSTQSEE286 pKa = 4.18NAKK289 pKa = 10.29QIVGVVSLPMSEE301 pKa = 4.41PPNVYY306 pKa = 10.35AKK308 pKa = 10.25EE309 pKa = 4.14AEE311 pKa = 4.32NIQAAIDD318 pKa = 3.46QRR320 pKa = 11.84ITRR323 pKa = 11.84KK324 pKa = 9.38QRR326 pKa = 11.84AFTANKK332 pKa = 9.02EE333 pKa = 4.12DD334 pKa = 4.41KK335 pKa = 10.33ILLGRR340 pKa = 11.84LVSEE344 pKa = 5.78AIGNDD349 pKa = 3.06PRR351 pKa = 11.84RR352 pKa = 11.84SLFSTRR358 pKa = 11.84RR359 pKa = 11.84VTEE362 pKa = 3.25WWEE365 pKa = 3.66THH367 pKa = 6.25LFGDD371 pKa = 4.67LKK373 pKa = 10.58SGKK376 pKa = 6.64WTEE379 pKa = 3.93EE380 pKa = 3.69RR381 pKa = 11.84LSRR384 pKa = 11.84TIEE387 pKa = 4.09GLCQRR392 pKa = 11.84INPKK396 pKa = 10.28FKK398 pKa = 10.58LSCDD402 pKa = 3.16IKK404 pKa = 10.84LEE406 pKa = 3.94PMPEE410 pKa = 4.1GKK412 pKa = 9.46PPRR415 pKa = 11.84MLIADD420 pKa = 3.77GDD422 pKa = 4.09EE423 pKa = 4.48GQVLALLTICCIEE436 pKa = 4.3DD437 pKa = 4.39LIKK440 pKa = 10.83KK441 pKa = 8.76HH442 pKa = 6.07LPKK445 pKa = 9.82KK446 pKa = 8.18TIKK449 pKa = 10.87GLGKK453 pKa = 8.5RR454 pKa = 11.84QAMEE458 pKa = 3.83RR459 pKa = 11.84VAQEE463 pKa = 3.57LRR465 pKa = 11.84APKK468 pKa = 9.86AAYY471 pKa = 10.0AKK473 pKa = 9.23TRR475 pKa = 11.84SAAKK479 pKa = 9.67SGQYY483 pKa = 9.7TGFSKK488 pKa = 10.1MVPPGVTVFEE498 pKa = 4.86GDD500 pKa = 3.03GSAWDD505 pKa = 4.23TTCSASLRR513 pKa = 11.84DD514 pKa = 3.86CVEE517 pKa = 3.82NPVIVHH523 pKa = 5.77VASILKK529 pKa = 9.26VHH531 pKa = 6.18MVQPEE536 pKa = 4.36GWVDD540 pKa = 3.14AHH542 pKa = 5.79TEE544 pKa = 3.58ISKK547 pKa = 9.34MEE549 pKa = 4.01KK550 pKa = 10.46LLIIFKK556 pKa = 10.87KK557 pKa = 9.99NGEE560 pKa = 3.94FRR562 pKa = 11.84KK563 pKa = 10.54YY564 pKa = 10.26IIDD567 pKa = 4.74AIRR570 pKa = 11.84RR571 pKa = 11.84SGHH574 pKa = 6.55RR575 pKa = 11.84GTSCLNWWTNFVCWHH590 pKa = 6.01CAIFEE595 pKa = 4.16KK596 pKa = 10.85PEE598 pKa = 3.9IFLDD602 pKa = 3.39PDD604 pKa = 2.99VRR606 pKa = 11.84YY607 pKa = 10.02GLDD610 pKa = 3.12HH611 pKa = 6.55SGVWRR616 pKa = 11.84WIATAFEE623 pKa = 4.82GDD625 pKa = 3.87DD626 pKa = 4.63SILSTTPRR634 pKa = 11.84IDD636 pKa = 3.42VKK638 pKa = 10.95SEE640 pKa = 3.64IYY642 pKa = 10.42VSIMQRR648 pKa = 11.84WEE650 pKa = 3.59RR651 pKa = 11.84LGFNMEE657 pKa = 3.45IFIRR661 pKa = 11.84EE662 pKa = 3.78EE663 pKa = 3.6RR664 pKa = 11.84AKK666 pKa = 10.02FTGYY670 pKa = 11.19YY671 pKa = 8.32MALDD675 pKa = 3.76NDD677 pKa = 4.11GTTGVLMPEE686 pKa = 4.11VDD688 pKa = 3.17RR689 pKa = 11.84CFARR693 pKa = 11.84SGVSCSPSMIEE704 pKa = 4.42CFKK707 pKa = 11.21KK708 pKa = 10.34EE709 pKa = 4.35DD710 pKa = 3.53RR711 pKa = 11.84TGCQSISRR719 pKa = 11.84AAALSRR725 pKa = 11.84AYY727 pKa = 9.65EE728 pKa = 4.03FAGLSPTISTKK739 pKa = 7.95YY740 pKa = 9.87LRR742 pKa = 11.84FYY744 pKa = 11.19EE745 pKa = 4.3SMSVSTKK752 pKa = 9.47VDD754 pKa = 3.28RR755 pKa = 11.84DD756 pKa = 3.25LKK758 pKa = 10.65MRR760 pKa = 11.84TCGGDD765 pKa = 3.12AEE767 pKa = 4.45FSEE770 pKa = 4.82KK771 pKa = 10.7EE772 pKa = 3.65IVAEE776 pKa = 4.1INLKK780 pKa = 10.7NGGAMSFDD788 pKa = 3.14SSEE791 pKa = 4.29RR792 pKa = 11.84DD793 pKa = 2.93RR794 pKa = 11.84LAAVGFKK801 pKa = 9.67CTEE804 pKa = 3.92EE805 pKa = 3.99EE806 pKa = 4.07LSRR809 pKa = 11.84FALRR813 pKa = 11.84MWDD816 pKa = 3.6YY817 pKa = 11.89DD818 pKa = 5.03LLKK821 pKa = 10.78DD822 pKa = 3.12WDD824 pKa = 4.1GFRR827 pKa = 11.84EE828 pKa = 4.13SLPEE832 pKa = 3.87SWRR835 pKa = 11.84MAA837 pKa = 3.76
MM1 pKa = 7.49TLPLSVANTTSLRR14 pKa = 11.84DD15 pKa = 3.27IEE17 pKa = 4.42GWMFKK22 pKa = 10.04IASEE26 pKa = 4.26RR27 pKa = 11.84PGMYY31 pKa = 9.49TIRR34 pKa = 11.84VRR36 pKa = 11.84LPQRR40 pKa = 11.84RR41 pKa = 11.84CGSCIKK47 pKa = 9.75LTRR50 pKa = 11.84SPLDD54 pKa = 3.56DD55 pKa = 3.54MEE57 pKa = 4.35EE58 pKa = 4.36VVVHH62 pKa = 5.08ITGEE66 pKa = 4.13EE67 pKa = 3.86FAKK70 pKa = 10.25LDD72 pKa = 3.83KK73 pKa = 11.27VIMNQSYY80 pKa = 10.49SEE82 pKa = 4.38SKK84 pKa = 10.48PKK86 pKa = 10.1VQAAIIYY93 pKa = 7.7QHH95 pKa = 7.13ASTIWPSHH103 pKa = 5.74TPDD106 pKa = 3.84DD107 pKa = 3.72VTMRR111 pKa = 11.84TLGAWAITQYY121 pKa = 10.67RR122 pKa = 11.84CNGGWAKK129 pKa = 10.66RR130 pKa = 11.84LLKK133 pKa = 10.3CIPCLQYY140 pKa = 9.57MQKK143 pKa = 10.16EE144 pKa = 4.11VHH146 pKa = 6.71LDD148 pKa = 3.57LKK150 pKa = 9.91GLRR153 pKa = 11.84MPNRR157 pKa = 11.84PVPPTPVEE165 pKa = 4.24SEE167 pKa = 4.01EE168 pKa = 4.26SSAGGTNGSTVRR180 pKa = 11.84TNTTATTAEE189 pKa = 4.82LSSQPTVEE197 pKa = 4.25SLPTITEE204 pKa = 3.97HH205 pKa = 8.08SDD207 pKa = 3.46EE208 pKa = 4.23EE209 pKa = 4.7DD210 pKa = 3.17EE211 pKa = 4.22NARR214 pKa = 11.84ANEE217 pKa = 4.28TPGEE221 pKa = 3.88AWARR225 pKa = 11.84WLAAGPPRR233 pKa = 11.84NEE235 pKa = 3.32IGYY238 pKa = 9.76VEE240 pKa = 4.87LGHH243 pKa = 6.84GNEE246 pKa = 4.75DD247 pKa = 3.61LHH249 pKa = 7.35PLGGASAEE257 pKa = 4.16DD258 pKa = 3.36SAIRR262 pKa = 11.84AEE264 pKa = 4.18RR265 pKa = 11.84QDD267 pKa = 3.48RR268 pKa = 11.84VLEE271 pKa = 4.04NAAGVGVVGQSTQSEE286 pKa = 4.18NAKK289 pKa = 10.29QIVGVVSLPMSEE301 pKa = 4.41PPNVYY306 pKa = 10.35AKK308 pKa = 10.25EE309 pKa = 4.14AEE311 pKa = 4.32NIQAAIDD318 pKa = 3.46QRR320 pKa = 11.84ITRR323 pKa = 11.84KK324 pKa = 9.38QRR326 pKa = 11.84AFTANKK332 pKa = 9.02EE333 pKa = 4.12DD334 pKa = 4.41KK335 pKa = 10.33ILLGRR340 pKa = 11.84LVSEE344 pKa = 5.78AIGNDD349 pKa = 3.06PRR351 pKa = 11.84RR352 pKa = 11.84SLFSTRR358 pKa = 11.84RR359 pKa = 11.84VTEE362 pKa = 3.25WWEE365 pKa = 3.66THH367 pKa = 6.25LFGDD371 pKa = 4.67LKK373 pKa = 10.58SGKK376 pKa = 6.64WTEE379 pKa = 3.93EE380 pKa = 3.69RR381 pKa = 11.84LSRR384 pKa = 11.84TIEE387 pKa = 4.09GLCQRR392 pKa = 11.84INPKK396 pKa = 10.28FKK398 pKa = 10.58LSCDD402 pKa = 3.16IKK404 pKa = 10.84LEE406 pKa = 3.94PMPEE410 pKa = 4.1GKK412 pKa = 9.46PPRR415 pKa = 11.84MLIADD420 pKa = 3.77GDD422 pKa = 4.09EE423 pKa = 4.48GQVLALLTICCIEE436 pKa = 4.3DD437 pKa = 4.39LIKK440 pKa = 10.83KK441 pKa = 8.76HH442 pKa = 6.07LPKK445 pKa = 9.82KK446 pKa = 8.18TIKK449 pKa = 10.87GLGKK453 pKa = 8.5RR454 pKa = 11.84QAMEE458 pKa = 3.83RR459 pKa = 11.84VAQEE463 pKa = 3.57LRR465 pKa = 11.84APKK468 pKa = 9.86AAYY471 pKa = 10.0AKK473 pKa = 9.23TRR475 pKa = 11.84SAAKK479 pKa = 9.67SGQYY483 pKa = 9.7TGFSKK488 pKa = 10.1MVPPGVTVFEE498 pKa = 4.86GDD500 pKa = 3.03GSAWDD505 pKa = 4.23TTCSASLRR513 pKa = 11.84DD514 pKa = 3.86CVEE517 pKa = 3.82NPVIVHH523 pKa = 5.77VASILKK529 pKa = 9.26VHH531 pKa = 6.18MVQPEE536 pKa = 4.36GWVDD540 pKa = 3.14AHH542 pKa = 5.79TEE544 pKa = 3.58ISKK547 pKa = 9.34MEE549 pKa = 4.01KK550 pKa = 10.46LLIIFKK556 pKa = 10.87KK557 pKa = 9.99NGEE560 pKa = 3.94FRR562 pKa = 11.84KK563 pKa = 10.54YY564 pKa = 10.26IIDD567 pKa = 4.74AIRR570 pKa = 11.84RR571 pKa = 11.84SGHH574 pKa = 6.55RR575 pKa = 11.84GTSCLNWWTNFVCWHH590 pKa = 6.01CAIFEE595 pKa = 4.16KK596 pKa = 10.85PEE598 pKa = 3.9IFLDD602 pKa = 3.39PDD604 pKa = 2.99VRR606 pKa = 11.84YY607 pKa = 10.02GLDD610 pKa = 3.12HH611 pKa = 6.55SGVWRR616 pKa = 11.84WIATAFEE623 pKa = 4.82GDD625 pKa = 3.87DD626 pKa = 4.63SILSTTPRR634 pKa = 11.84IDD636 pKa = 3.42VKK638 pKa = 10.95SEE640 pKa = 3.64IYY642 pKa = 10.42VSIMQRR648 pKa = 11.84WEE650 pKa = 3.59RR651 pKa = 11.84LGFNMEE657 pKa = 3.45IFIRR661 pKa = 11.84EE662 pKa = 3.78EE663 pKa = 3.6RR664 pKa = 11.84AKK666 pKa = 10.02FTGYY670 pKa = 11.19YY671 pKa = 8.32MALDD675 pKa = 3.76NDD677 pKa = 4.11GTTGVLMPEE686 pKa = 4.11VDD688 pKa = 3.17RR689 pKa = 11.84CFARR693 pKa = 11.84SGVSCSPSMIEE704 pKa = 4.42CFKK707 pKa = 11.21KK708 pKa = 10.34EE709 pKa = 4.35DD710 pKa = 3.53RR711 pKa = 11.84TGCQSISRR719 pKa = 11.84AAALSRR725 pKa = 11.84AYY727 pKa = 9.65EE728 pKa = 4.03FAGLSPTISTKK739 pKa = 7.95YY740 pKa = 9.87LRR742 pKa = 11.84FYY744 pKa = 11.19EE745 pKa = 4.3SMSVSTKK752 pKa = 9.47VDD754 pKa = 3.28RR755 pKa = 11.84DD756 pKa = 3.25LKK758 pKa = 10.65MRR760 pKa = 11.84TCGGDD765 pKa = 3.12AEE767 pKa = 4.45FSEE770 pKa = 4.82KK771 pKa = 10.7EE772 pKa = 3.65IVAEE776 pKa = 4.1INLKK780 pKa = 10.7NGGAMSFDD788 pKa = 3.14SSEE791 pKa = 4.29RR792 pKa = 11.84DD793 pKa = 2.93RR794 pKa = 11.84LAAVGFKK801 pKa = 9.67CTEE804 pKa = 3.92EE805 pKa = 3.99EE806 pKa = 4.07LSRR809 pKa = 11.84FALRR813 pKa = 11.84MWDD816 pKa = 3.6YY817 pKa = 11.89DD818 pKa = 5.03LLKK821 pKa = 10.78DD822 pKa = 3.12WDD824 pKa = 4.1GFRR827 pKa = 11.84EE828 pKa = 4.13SLPEE832 pKa = 3.87SWRR835 pKa = 11.84MAA837 pKa = 3.76
Molecular weight: 93.96 kDa
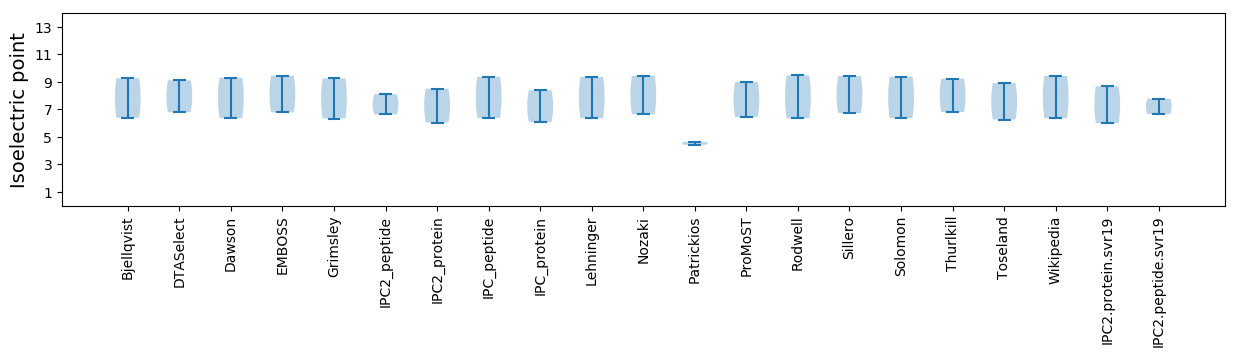
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KLA9|A0A1L3KLA9_9VIRU RNA-dependent RNA polymerase OS=Beihai weivirus-like virus 9 OX=1922756 PE=4 SV=1
MM1 pKa = 7.04VLHH4 pKa = 7.1RR5 pKa = 11.84GKK7 pKa = 10.23KK8 pKa = 8.68HH9 pKa = 5.47AALGRR14 pKa = 11.84KK15 pKa = 6.15TAKK18 pKa = 9.0THH20 pKa = 4.69TKK22 pKa = 8.67YY23 pKa = 9.77ATRR26 pKa = 11.84ARR28 pKa = 11.84GVVSAAPPLTKK39 pKa = 10.57GGIARR44 pKa = 11.84HH45 pKa = 4.96KK46 pKa = 10.57AEE48 pKa = 3.79RR49 pKa = 11.84HH50 pKa = 4.79KK51 pKa = 11.13VISGYY56 pKa = 9.81RR57 pKa = 11.84RR58 pKa = 11.84EE59 pKa = 3.98EE60 pKa = 4.08DD61 pKa = 3.37KK62 pKa = 11.61LNAFLPGHH70 pKa = 6.38LTLPRR75 pKa = 11.84AVAPYY80 pKa = 9.83SVVRR84 pKa = 11.84TIVNFPLACSDD95 pKa = 3.64RR96 pKa = 11.84GKK98 pKa = 9.51MCVIQPMVNEE108 pKa = 4.41DD109 pKa = 3.21AGSNKK114 pKa = 8.32ITFTNCIGWVSDD126 pKa = 3.58TDD128 pKa = 3.68QSSGPVPTTNTPLGINMPSVGNTGVEE154 pKa = 4.32CVPAAISVRR163 pKa = 11.84ITCPSPLIAATGQLFLGRR181 pKa = 11.84WNMPADD187 pKa = 3.86RR188 pKa = 11.84KK189 pKa = 11.02NYY191 pKa = 8.64VHH193 pKa = 6.89YY194 pKa = 11.13DD195 pKa = 3.48DD196 pKa = 4.69MEE198 pKa = 5.72KK199 pKa = 11.17GFLAFAKK206 pKa = 9.96PEE208 pKa = 4.4PYY210 pKa = 8.75TAAMMATCTRR220 pKa = 11.84QVSAIPRR227 pKa = 11.84DD228 pKa = 3.7FNDD231 pKa = 3.01YY232 pKa = 11.39CEE234 pKa = 4.03FHH236 pKa = 7.34NYY238 pKa = 9.45EE239 pKa = 4.37DD240 pKa = 4.2TNFTDD245 pKa = 5.26ALKK248 pKa = 9.89PWGGMTPILYY258 pKa = 10.07CLSPVSGGNVTQYY271 pKa = 11.54NLQICVEE278 pKa = 3.49WRR280 pKa = 11.84YY281 pKa = 9.8RR282 pKa = 11.84FKK284 pKa = 10.57MDD286 pKa = 3.98DD287 pKa = 3.54PAASSHH293 pKa = 5.82EE294 pKa = 3.93FHH296 pKa = 7.08FPHH299 pKa = 7.53PIDD302 pKa = 3.98KK303 pKa = 11.05VNGLIARR310 pKa = 11.84MSGKK314 pKa = 9.82PGVDD318 pKa = 3.25VVKK321 pKa = 10.68KK322 pKa = 10.39GDD324 pKa = 3.56KK325 pKa = 10.59SPPDD329 pKa = 3.78VLTKK333 pKa = 10.83
MM1 pKa = 7.04VLHH4 pKa = 7.1RR5 pKa = 11.84GKK7 pKa = 10.23KK8 pKa = 8.68HH9 pKa = 5.47AALGRR14 pKa = 11.84KK15 pKa = 6.15TAKK18 pKa = 9.0THH20 pKa = 4.69TKK22 pKa = 8.67YY23 pKa = 9.77ATRR26 pKa = 11.84ARR28 pKa = 11.84GVVSAAPPLTKK39 pKa = 10.57GGIARR44 pKa = 11.84HH45 pKa = 4.96KK46 pKa = 10.57AEE48 pKa = 3.79RR49 pKa = 11.84HH50 pKa = 4.79KK51 pKa = 11.13VISGYY56 pKa = 9.81RR57 pKa = 11.84RR58 pKa = 11.84EE59 pKa = 3.98EE60 pKa = 4.08DD61 pKa = 3.37KK62 pKa = 11.61LNAFLPGHH70 pKa = 6.38LTLPRR75 pKa = 11.84AVAPYY80 pKa = 9.83SVVRR84 pKa = 11.84TIVNFPLACSDD95 pKa = 3.64RR96 pKa = 11.84GKK98 pKa = 9.51MCVIQPMVNEE108 pKa = 4.41DD109 pKa = 3.21AGSNKK114 pKa = 8.32ITFTNCIGWVSDD126 pKa = 3.58TDD128 pKa = 3.68QSSGPVPTTNTPLGINMPSVGNTGVEE154 pKa = 4.32CVPAAISVRR163 pKa = 11.84ITCPSPLIAATGQLFLGRR181 pKa = 11.84WNMPADD187 pKa = 3.86RR188 pKa = 11.84KK189 pKa = 11.02NYY191 pKa = 8.64VHH193 pKa = 6.89YY194 pKa = 11.13DD195 pKa = 3.48DD196 pKa = 4.69MEE198 pKa = 5.72KK199 pKa = 11.17GFLAFAKK206 pKa = 9.96PEE208 pKa = 4.4PYY210 pKa = 8.75TAAMMATCTRR220 pKa = 11.84QVSAIPRR227 pKa = 11.84DD228 pKa = 3.7FNDD231 pKa = 3.01YY232 pKa = 11.39CEE234 pKa = 4.03FHH236 pKa = 7.34NYY238 pKa = 9.45EE239 pKa = 4.37DD240 pKa = 4.2TNFTDD245 pKa = 5.26ALKK248 pKa = 9.89PWGGMTPILYY258 pKa = 10.07CLSPVSGGNVTQYY271 pKa = 11.54NLQICVEE278 pKa = 3.49WRR280 pKa = 11.84YY281 pKa = 9.8RR282 pKa = 11.84FKK284 pKa = 10.57MDD286 pKa = 3.98DD287 pKa = 3.54PAASSHH293 pKa = 5.82EE294 pKa = 3.93FHH296 pKa = 7.08FPHH299 pKa = 7.53PIDD302 pKa = 3.98KK303 pKa = 11.05VNGLIARR310 pKa = 11.84MSGKK314 pKa = 9.82PGVDD318 pKa = 3.25VVKK321 pKa = 10.68KK322 pKa = 10.39GDD324 pKa = 3.56KK325 pKa = 10.59SPPDD329 pKa = 3.78VLTKK333 pKa = 10.83
Molecular weight: 36.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1170 |
333 |
837 |
585.0 |
65.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.034 ± 0.371 | 2.479 ± 0.123 |
5.299 ± 0.058 | 7.009 ± 2.036 |
3.248 ± 0.195 | 7.009 ± 0.274 |
2.222 ± 0.594 | 5.556 ± 0.578 |
6.068 ± 0.296 | 6.923 ± 0.669 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.077 ± 0.124 | 3.504 ± 0.715 |
5.812 ± 1.262 | 2.479 ± 0.372 |
6.838 ± 0.622 | 7.009 ± 0.881 |
6.581 ± 0.344 | 6.325 ± 0.815 |
2.051 ± 0.467 | 2.479 ± 0.453 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
