
Odonata-associated circular virus-17
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.7
Get precalculated fractions of proteins
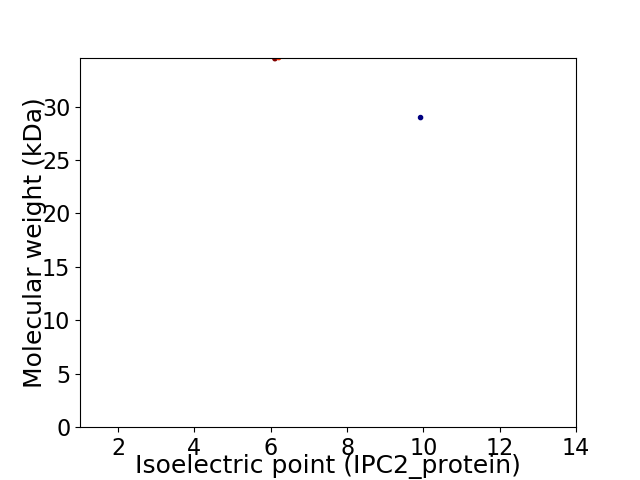
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
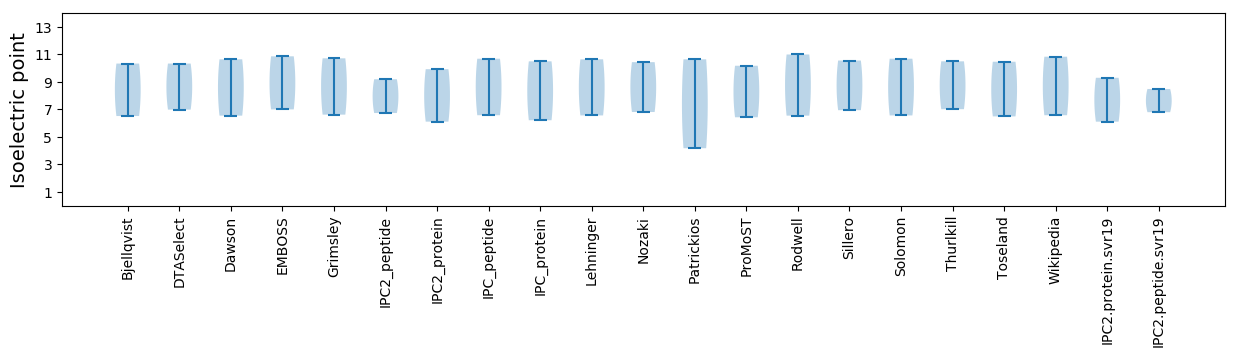
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4UI13|A0A0B4UI13_9VIRU Replication-associated protein OS=Odonata-associated circular virus-17 OX=1592117 PE=4 SV=1
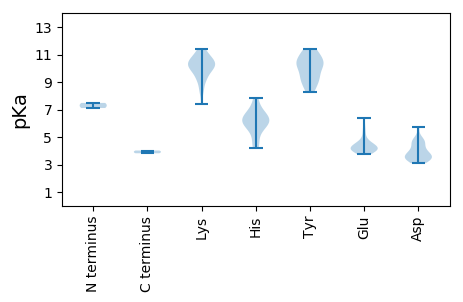
MM1 pKa = 7.11TSEE4 pKa = 4.34VMTSIAPGNTKK15 pKa = 9.8PVALKK20 pKa = 10.36ARR22 pKa = 11.84AYY24 pKa = 10.51QFTLNQPDD32 pKa = 4.27VYY34 pKa = 11.33DD35 pKa = 3.54KK36 pKa = 11.34LKK38 pKa = 11.21NILTGLKK45 pKa = 9.66TMDD48 pKa = 3.55YY49 pKa = 10.4LISCRR54 pKa = 11.84EE55 pKa = 3.93TAPSTGHH62 pKa = 4.66VHH64 pKa = 4.23MHH66 pKa = 5.87VYY68 pKa = 10.84AHH70 pKa = 7.03FSQQYY75 pKa = 9.48RR76 pKa = 11.84LPKK79 pKa = 9.93KK80 pKa = 10.29IRR82 pKa = 11.84DD83 pKa = 3.86LGCHH87 pKa = 6.22IEE89 pKa = 4.0ICLVGAKK96 pKa = 9.75QNIAYY101 pKa = 8.87IRR103 pKa = 11.84KK104 pKa = 9.17GGDD107 pKa = 3.1IIDD110 pKa = 4.77EE111 pKa = 4.6IGDD114 pKa = 3.83EE115 pKa = 4.07PHH117 pKa = 6.09QGARR121 pKa = 11.84TVGEE125 pKa = 4.31LMEE128 pKa = 6.42ADD130 pKa = 3.53IKK132 pKa = 11.43SIDD135 pKa = 3.32PHH137 pKa = 7.82LYY139 pKa = 10.06RR140 pKa = 11.84IKK142 pKa = 10.63KK143 pKa = 10.07QIDD146 pKa = 3.59AEE148 pKa = 4.21QHH150 pKa = 6.37DD151 pKa = 5.7LDD153 pKa = 4.59TFMSMLDD160 pKa = 3.77EE161 pKa = 5.75IEE163 pKa = 5.36ADD165 pKa = 3.72DD166 pKa = 4.86LKK168 pKa = 11.22APTVIYY174 pKa = 9.35ITGGTGKK181 pKa = 10.52GKK183 pKa = 9.0TYY185 pKa = 10.53SAYY188 pKa = 9.67KK189 pKa = 10.01RR190 pKa = 11.84ALKK193 pKa = 9.92EE194 pKa = 3.89YY195 pKa = 8.86PKK197 pKa = 10.86KK198 pKa = 10.86KK199 pKa = 9.65IGKK202 pKa = 8.31LTLKK206 pKa = 10.49NDD208 pKa = 4.65FIDD211 pKa = 4.76IVNGDD216 pKa = 3.48NAEE219 pKa = 4.15CFVIEE224 pKa = 4.5EE225 pKa = 4.39FRR227 pKa = 11.84PSQIKK232 pKa = 10.58ASDD235 pKa = 3.47FLQLTDD241 pKa = 3.67KK242 pKa = 11.17YY243 pKa = 11.22GYY245 pKa = 9.65RR246 pKa = 11.84CNIKK250 pKa = 10.62GGFATLRR257 pKa = 11.84PKK259 pKa = 10.17MIIICSIIQPEE270 pKa = 4.47HH271 pKa = 6.28IYY273 pKa = 10.7KK274 pKa = 10.81EE275 pKa = 4.23EE276 pKa = 4.02VNKK279 pKa = 10.37QFLRR283 pKa = 11.84RR284 pKa = 11.84ITEE287 pKa = 4.47RR288 pKa = 11.84IDD290 pKa = 3.48LGPEE294 pKa = 4.16LDD296 pKa = 4.75PVDD299 pKa = 4.38EE300 pKa = 5.0LYY302 pKa = 10.78EE303 pKa = 4.02
MM1 pKa = 7.11TSEE4 pKa = 4.34VMTSIAPGNTKK15 pKa = 9.8PVALKK20 pKa = 10.36ARR22 pKa = 11.84AYY24 pKa = 10.51QFTLNQPDD32 pKa = 4.27VYY34 pKa = 11.33DD35 pKa = 3.54KK36 pKa = 11.34LKK38 pKa = 11.21NILTGLKK45 pKa = 9.66TMDD48 pKa = 3.55YY49 pKa = 10.4LISCRR54 pKa = 11.84EE55 pKa = 3.93TAPSTGHH62 pKa = 4.66VHH64 pKa = 4.23MHH66 pKa = 5.87VYY68 pKa = 10.84AHH70 pKa = 7.03FSQQYY75 pKa = 9.48RR76 pKa = 11.84LPKK79 pKa = 9.93KK80 pKa = 10.29IRR82 pKa = 11.84DD83 pKa = 3.86LGCHH87 pKa = 6.22IEE89 pKa = 4.0ICLVGAKK96 pKa = 9.75QNIAYY101 pKa = 8.87IRR103 pKa = 11.84KK104 pKa = 9.17GGDD107 pKa = 3.1IIDD110 pKa = 4.77EE111 pKa = 4.6IGDD114 pKa = 3.83EE115 pKa = 4.07PHH117 pKa = 6.09QGARR121 pKa = 11.84TVGEE125 pKa = 4.31LMEE128 pKa = 6.42ADD130 pKa = 3.53IKK132 pKa = 11.43SIDD135 pKa = 3.32PHH137 pKa = 7.82LYY139 pKa = 10.06RR140 pKa = 11.84IKK142 pKa = 10.63KK143 pKa = 10.07QIDD146 pKa = 3.59AEE148 pKa = 4.21QHH150 pKa = 6.37DD151 pKa = 5.7LDD153 pKa = 4.59TFMSMLDD160 pKa = 3.77EE161 pKa = 5.75IEE163 pKa = 5.36ADD165 pKa = 3.72DD166 pKa = 4.86LKK168 pKa = 11.22APTVIYY174 pKa = 9.35ITGGTGKK181 pKa = 10.52GKK183 pKa = 9.0TYY185 pKa = 10.53SAYY188 pKa = 9.67KK189 pKa = 10.01RR190 pKa = 11.84ALKK193 pKa = 9.92EE194 pKa = 3.89YY195 pKa = 8.86PKK197 pKa = 10.86KK198 pKa = 10.86KK199 pKa = 9.65IGKK202 pKa = 8.31LTLKK206 pKa = 10.49NDD208 pKa = 4.65FIDD211 pKa = 4.76IVNGDD216 pKa = 3.48NAEE219 pKa = 4.15CFVIEE224 pKa = 4.5EE225 pKa = 4.39FRR227 pKa = 11.84PSQIKK232 pKa = 10.58ASDD235 pKa = 3.47FLQLTDD241 pKa = 3.67KK242 pKa = 11.17YY243 pKa = 11.22GYY245 pKa = 9.65RR246 pKa = 11.84CNIKK250 pKa = 10.62GGFATLRR257 pKa = 11.84PKK259 pKa = 10.17MIIICSIIQPEE270 pKa = 4.47HH271 pKa = 6.28IYY273 pKa = 10.7KK274 pKa = 10.81EE275 pKa = 4.23EE276 pKa = 4.02VNKK279 pKa = 10.37QFLRR283 pKa = 11.84RR284 pKa = 11.84ITEE287 pKa = 4.47RR288 pKa = 11.84IDD290 pKa = 3.48LGPEE294 pKa = 4.16LDD296 pKa = 4.75PVDD299 pKa = 4.38EE300 pKa = 5.0LYY302 pKa = 10.78EE303 pKa = 4.02
Molecular weight: 34.48 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UI13|A0A0B4UI13_9VIRU Replication-associated protein OS=Odonata-associated circular virus-17 OX=1592117 PE=4 SV=1
MM1 pKa = 7.48VIFLEE6 pKa = 4.67PSFSLVKK13 pKa = 10.36MSQTQIVDD21 pKa = 3.4YY22 pKa = 10.94VFYY25 pKa = 10.23SCYY28 pKa = 8.42MAYY31 pKa = 10.26GRR33 pKa = 11.84RR34 pKa = 11.84STYY37 pKa = 9.23RR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 10.06GRR42 pKa = 11.84RR43 pKa = 11.84GNRR46 pKa = 11.84SLTTRR51 pKa = 11.84RR52 pKa = 11.84IFNNKK57 pKa = 7.4GAKK60 pKa = 9.28AQAKK64 pKa = 9.37QIYY67 pKa = 8.6ALKK70 pKa = 10.14RR71 pKa = 11.84AVNRR75 pKa = 11.84VRR77 pKa = 11.84AQCKK81 pKa = 10.13PEE83 pKa = 3.77VKK85 pKa = 9.92YY86 pKa = 11.23VKK88 pKa = 10.54SEE90 pKa = 4.0TQNRR94 pKa = 11.84ALGLEE99 pKa = 4.1HH100 pKa = 6.68QGASFLQYY108 pKa = 10.21YY109 pKa = 9.22AQLPMPGISAGTGDD123 pKa = 4.22GNRR126 pKa = 11.84IGDD129 pKa = 4.03NVRR132 pKa = 11.84LLPMKK137 pKa = 10.5LGINIRR143 pKa = 11.84YY144 pKa = 8.25EE145 pKa = 4.0EE146 pKa = 4.27YY147 pKa = 10.23MNSLTKK153 pKa = 10.09TYY155 pKa = 10.39PVDD158 pKa = 3.22IPLNSTGGQVRR169 pKa = 11.84IIAVQAITAHH179 pKa = 5.97SNEE182 pKa = 4.15PQLVDD187 pKa = 3.94LMNANEE193 pKa = 4.09YY194 pKa = 11.43NMGAIGAAGMMNMSFKK210 pKa = 10.73RR211 pKa = 11.84GITSHH216 pKa = 6.4YY217 pKa = 9.17KK218 pKa = 9.22ILMNKK223 pKa = 8.42VVTVSKK229 pKa = 10.61DD230 pKa = 3.21RR231 pKa = 11.84PIVNKK236 pKa = 10.23RR237 pKa = 11.84YY238 pKa = 9.95LIRR241 pKa = 11.84PAIKK245 pKa = 9.16HH246 pKa = 5.6LKK248 pKa = 8.42VGRR251 pKa = 11.84RR252 pKa = 11.84TDD254 pKa = 3.27STT256 pKa = 3.82
MM1 pKa = 7.48VIFLEE6 pKa = 4.67PSFSLVKK13 pKa = 10.36MSQTQIVDD21 pKa = 3.4YY22 pKa = 10.94VFYY25 pKa = 10.23SCYY28 pKa = 8.42MAYY31 pKa = 10.26GRR33 pKa = 11.84RR34 pKa = 11.84STYY37 pKa = 9.23RR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 10.06GRR42 pKa = 11.84RR43 pKa = 11.84GNRR46 pKa = 11.84SLTTRR51 pKa = 11.84RR52 pKa = 11.84IFNNKK57 pKa = 7.4GAKK60 pKa = 9.28AQAKK64 pKa = 9.37QIYY67 pKa = 8.6ALKK70 pKa = 10.14RR71 pKa = 11.84AVNRR75 pKa = 11.84VRR77 pKa = 11.84AQCKK81 pKa = 10.13PEE83 pKa = 3.77VKK85 pKa = 9.92YY86 pKa = 11.23VKK88 pKa = 10.54SEE90 pKa = 4.0TQNRR94 pKa = 11.84ALGLEE99 pKa = 4.1HH100 pKa = 6.68QGASFLQYY108 pKa = 10.21YY109 pKa = 9.22AQLPMPGISAGTGDD123 pKa = 4.22GNRR126 pKa = 11.84IGDD129 pKa = 4.03NVRR132 pKa = 11.84LLPMKK137 pKa = 10.5LGINIRR143 pKa = 11.84YY144 pKa = 8.25EE145 pKa = 4.0EE146 pKa = 4.27YY147 pKa = 10.23MNSLTKK153 pKa = 10.09TYY155 pKa = 10.39PVDD158 pKa = 3.22IPLNSTGGQVRR169 pKa = 11.84IIAVQAITAHH179 pKa = 5.97SNEE182 pKa = 4.15PQLVDD187 pKa = 3.94LMNANEE193 pKa = 4.09YY194 pKa = 11.43NMGAIGAAGMMNMSFKK210 pKa = 10.73RR211 pKa = 11.84GITSHH216 pKa = 6.4YY217 pKa = 9.17KK218 pKa = 9.22ILMNKK223 pKa = 8.42VVTVSKK229 pKa = 10.61DD230 pKa = 3.21RR231 pKa = 11.84PIVNKK236 pKa = 10.23RR237 pKa = 11.84YY238 pKa = 9.95LIRR241 pKa = 11.84PAIKK245 pKa = 9.16HH246 pKa = 5.6LKK248 pKa = 8.42VGRR251 pKa = 11.84RR252 pKa = 11.84TDD254 pKa = 3.27STT256 pKa = 3.82
Molecular weight: 28.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
559 |
256 |
303 |
279.5 |
31.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.619 ± 0.533 | 1.431 ± 0.431 |
5.367 ± 1.748 | 5.188 ± 1.37 |
2.683 ± 0.225 | 6.977 ± 0.296 |
2.326 ± 0.507 | 8.945 ± 1.27 |
8.229 ± 0.795 | 7.513 ± 0.32 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.578 ± 0.737 | 4.651 ± 1.321 |
4.293 ± 0.257 | 4.293 ± 0.262 |
6.619 ± 1.571 | 4.83 ± 0.943 |
5.725 ± 0.17 | 5.367 ± 1.105 |
0.0 ± 0.0 | 5.367 ± 0.327 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
