
Arthrospira platensis (strain NIES-39 / IAM M-135) (Spirulina platensis)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Oscillatoriophycideae; Oscillatoriales; Microcoleaceae; Arthrospira; Arthrospira platensis
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
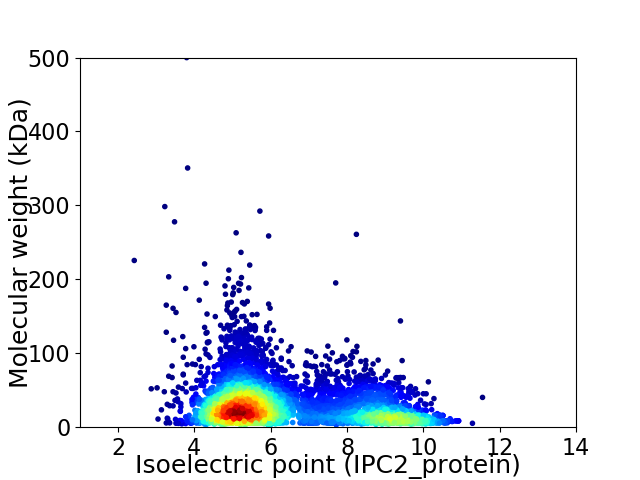
Virtual 2D-PAGE plot for 6009 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D4ZPZ7|D4ZPZ7_ARTPN Methyltransferase OS=Arthrospira platensis (strain NIES-39 / IAM M-135) OX=696747 GN=NIES39_Q01180 PE=3 SV=1
MM1 pKa = 6.9NTLNVAQPYY10 pKa = 7.75EE11 pKa = 4.24IPSNEE16 pKa = 3.52NDD18 pKa = 3.31VFLARR23 pKa = 11.84YY24 pKa = 8.71EE25 pKa = 4.15PDD27 pKa = 3.38GSQVWTTRR35 pKa = 11.84LASNRR40 pKa = 11.84SEE42 pKa = 3.85IAHH45 pKa = 6.84ALTTGLDD52 pKa = 3.38GSIYY56 pKa = 10.46VAGFTSGDD64 pKa = 3.25LGGEE68 pKa = 4.12INNGGTDD75 pKa = 3.52AFISNYY81 pKa = 9.92EE82 pKa = 3.8PDD84 pKa = 3.55GTLTRR89 pKa = 11.84TRR91 pKa = 11.84LLGTSNWDD99 pKa = 3.09GARR102 pKa = 11.84ALTTGFDD109 pKa = 3.48GSIYY113 pKa = 9.8MAGEE117 pKa = 3.73TWGDD121 pKa = 3.45LNGEE125 pKa = 4.32INQSGADD132 pKa = 3.7GFIAKK137 pKa = 9.74YY138 pKa = 10.07EE139 pKa = 4.12PDD141 pKa = 3.41GTLVWTRR148 pKa = 11.84LLGSNGWGAAHH159 pKa = 7.73ALTTGIDD166 pKa = 3.41GFIYY170 pKa = 10.6VAGLTEE176 pKa = 4.12GDD178 pKa = 3.29INEE181 pKa = 4.78EE182 pKa = 3.45IFNRR186 pKa = 11.84NRR188 pKa = 11.84GGNALISKK196 pKa = 8.73YY197 pKa = 9.65EE198 pKa = 4.14PEE200 pKa = 5.41DD201 pKa = 3.46GTLVWTTQLGTPNPDD216 pKa = 2.95EE217 pKa = 5.2ANALTTGLDD226 pKa = 3.35GSIYY230 pKa = 9.0MAGWTQGNLGGEE242 pKa = 4.32MNRR245 pKa = 11.84GYY247 pKa = 11.09SDD249 pKa = 3.75GFIAKK254 pKa = 10.04YY255 pKa = 8.38EE256 pKa = 4.16TDD258 pKa = 4.33GALAWTRR265 pKa = 11.84LLASNDD271 pKa = 2.96WDD273 pKa = 3.73GANALTTRR281 pKa = 11.84IDD283 pKa = 3.38GSIYY287 pKa = 9.09IAGYY291 pKa = 8.17TRR293 pKa = 11.84GNLNGEE299 pKa = 4.32INNGASDD306 pKa = 3.77AFMSRR311 pKa = 11.84VIVDD315 pKa = 3.48GTVAEE320 pKa = 4.39PVPPPEE326 pKa = 5.44FIDD329 pKa = 4.2PLLPLNGDD337 pKa = 4.42YY338 pKa = 10.83IPIAGDD344 pKa = 3.57FNGSGITDD352 pKa = 3.87ILWYY356 pKa = 10.61APGPDD361 pKa = 4.95PDD363 pKa = 4.21YY364 pKa = 10.37MWYY367 pKa = 9.75FHH369 pKa = 7.39ADD371 pKa = 2.86GSYY374 pKa = 10.3GSRR377 pKa = 11.84LFMINGYY384 pKa = 8.57YY385 pKa = 9.81IPIAGDD391 pKa = 3.61FNGSGITDD399 pKa = 3.87ILWYY403 pKa = 10.61APGPDD408 pKa = 4.87PDD410 pKa = 4.13FMWYY414 pKa = 10.32FNQDD418 pKa = 2.35GNYY421 pKa = 10.54DD422 pKa = 4.11GFQLTVDD429 pKa = 3.43GHH431 pKa = 5.41YY432 pKa = 11.13APVPADD438 pKa = 3.38LEE440 pKa = 4.66VFF442 pKa = 3.88
MM1 pKa = 6.9NTLNVAQPYY10 pKa = 7.75EE11 pKa = 4.24IPSNEE16 pKa = 3.52NDD18 pKa = 3.31VFLARR23 pKa = 11.84YY24 pKa = 8.71EE25 pKa = 4.15PDD27 pKa = 3.38GSQVWTTRR35 pKa = 11.84LASNRR40 pKa = 11.84SEE42 pKa = 3.85IAHH45 pKa = 6.84ALTTGLDD52 pKa = 3.38GSIYY56 pKa = 10.46VAGFTSGDD64 pKa = 3.25LGGEE68 pKa = 4.12INNGGTDD75 pKa = 3.52AFISNYY81 pKa = 9.92EE82 pKa = 3.8PDD84 pKa = 3.55GTLTRR89 pKa = 11.84TRR91 pKa = 11.84LLGTSNWDD99 pKa = 3.09GARR102 pKa = 11.84ALTTGFDD109 pKa = 3.48GSIYY113 pKa = 9.8MAGEE117 pKa = 3.73TWGDD121 pKa = 3.45LNGEE125 pKa = 4.32INQSGADD132 pKa = 3.7GFIAKK137 pKa = 9.74YY138 pKa = 10.07EE139 pKa = 4.12PDD141 pKa = 3.41GTLVWTRR148 pKa = 11.84LLGSNGWGAAHH159 pKa = 7.73ALTTGIDD166 pKa = 3.41GFIYY170 pKa = 10.6VAGLTEE176 pKa = 4.12GDD178 pKa = 3.29INEE181 pKa = 4.78EE182 pKa = 3.45IFNRR186 pKa = 11.84NRR188 pKa = 11.84GGNALISKK196 pKa = 8.73YY197 pKa = 9.65EE198 pKa = 4.14PEE200 pKa = 5.41DD201 pKa = 3.46GTLVWTTQLGTPNPDD216 pKa = 2.95EE217 pKa = 5.2ANALTTGLDD226 pKa = 3.35GSIYY230 pKa = 9.0MAGWTQGNLGGEE242 pKa = 4.32MNRR245 pKa = 11.84GYY247 pKa = 11.09SDD249 pKa = 3.75GFIAKK254 pKa = 10.04YY255 pKa = 8.38EE256 pKa = 4.16TDD258 pKa = 4.33GALAWTRR265 pKa = 11.84LLASNDD271 pKa = 2.96WDD273 pKa = 3.73GANALTTRR281 pKa = 11.84IDD283 pKa = 3.38GSIYY287 pKa = 9.09IAGYY291 pKa = 8.17TRR293 pKa = 11.84GNLNGEE299 pKa = 4.32INNGASDD306 pKa = 3.77AFMSRR311 pKa = 11.84VIVDD315 pKa = 3.48GTVAEE320 pKa = 4.39PVPPPEE326 pKa = 5.44FIDD329 pKa = 4.2PLLPLNGDD337 pKa = 4.42YY338 pKa = 10.83IPIAGDD344 pKa = 3.57FNGSGITDD352 pKa = 3.87ILWYY356 pKa = 10.61APGPDD361 pKa = 4.95PDD363 pKa = 4.21YY364 pKa = 10.37MWYY367 pKa = 9.75FHH369 pKa = 7.39ADD371 pKa = 2.86GSYY374 pKa = 10.3GSRR377 pKa = 11.84LFMINGYY384 pKa = 8.57YY385 pKa = 9.81IPIAGDD391 pKa = 3.61FNGSGITDD399 pKa = 3.87ILWYY403 pKa = 10.61APGPDD408 pKa = 4.87PDD410 pKa = 4.13FMWYY414 pKa = 10.32FNQDD418 pKa = 2.35GNYY421 pKa = 10.54DD422 pKa = 4.11GFQLTVDD429 pKa = 3.43GHH431 pKa = 5.41YY432 pKa = 11.13APVPADD438 pKa = 3.38LEE440 pKa = 4.66VFF442 pKa = 3.88
Molecular weight: 47.66 kDa
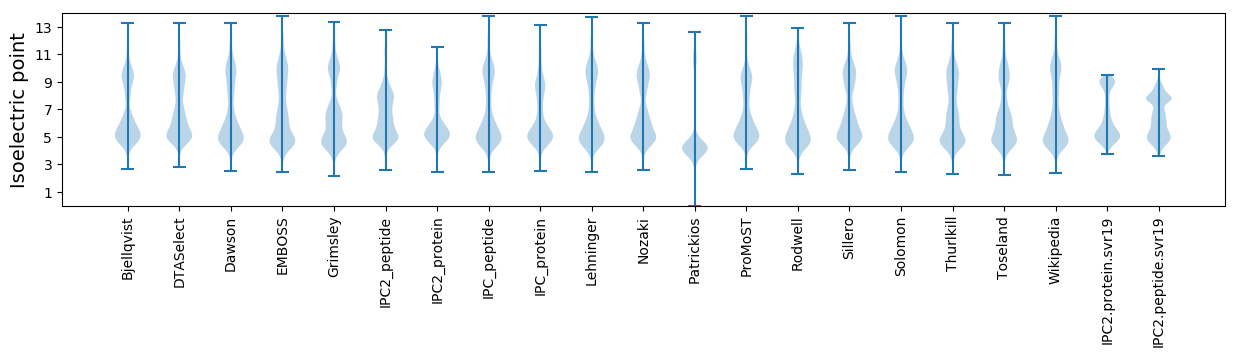
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D4ZQM7|D4ZQM7_ARTPN Reverse transcriptase OS=Arthrospira platensis (strain NIES-39 / IAM M-135) OX=696747 GN=NIES39_D07940 PE=4 SV=1
MM1 pKa = 7.42AQQTLKK7 pKa = 9.97GTSRR11 pKa = 11.84KK12 pKa = 9.14KK13 pKa = 10.39KK14 pKa = 8.02RR15 pKa = 11.84VSGFRR20 pKa = 11.84VRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84NGRR29 pKa = 11.84AVIRR33 pKa = 11.84SRR35 pKa = 11.84RR36 pKa = 11.84QKK38 pKa = 9.97GRR40 pKa = 11.84SRR42 pKa = 11.84LSVV45 pKa = 3.01
MM1 pKa = 7.42AQQTLKK7 pKa = 9.97GTSRR11 pKa = 11.84KK12 pKa = 9.14KK13 pKa = 10.39KK14 pKa = 8.02RR15 pKa = 11.84VSGFRR20 pKa = 11.84VRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84NGRR29 pKa = 11.84AVIRR33 pKa = 11.84SRR35 pKa = 11.84RR36 pKa = 11.84QKK38 pKa = 9.97GRR40 pKa = 11.84SRR42 pKa = 11.84LSVV45 pKa = 3.01
Molecular weight: 5.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1745523 |
29 |
4747 |
290.5 |
32.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.379 ± 0.034 | 1.064 ± 0.015 |
5.262 ± 0.033 | 6.288 ± 0.04 |
3.807 ± 0.025 | 6.836 ± 0.04 |
1.943 ± 0.017 | 6.901 ± 0.034 |
4.511 ± 0.041 | 10.539 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.984 ± 0.016 | 4.514 ± 0.031 |
5.062 ± 0.032 | 5.299 ± 0.035 |
5.494 ± 0.028 | 6.531 ± 0.058 |
5.652 ± 0.036 | 6.221 ± 0.033 |
1.565 ± 0.017 | 3.15 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
