
Hubei tombus-like virus 7
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.92
Get precalculated fractions of proteins
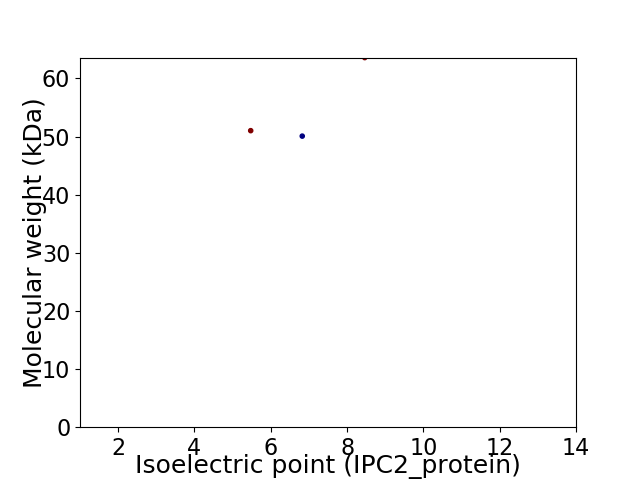
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
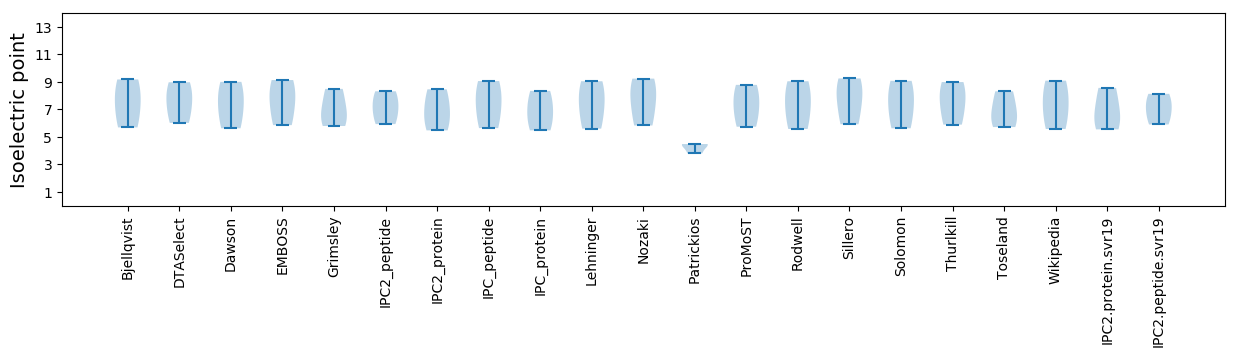
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGP4|A0A1L3KGP4_9VIRU RNA-directed RNA polymerase OS=Hubei tombus-like virus 7 OX=1923294 PE=4 SV=1
MM1 pKa = 7.35SFAYY5 pKa = 9.28TYY7 pKa = 9.72FFQPKK12 pKa = 9.23RR13 pKa = 11.84APTFDD18 pKa = 3.64FSTTRR23 pKa = 11.84RR24 pKa = 11.84LDD26 pKa = 3.36AFNPATALTGGPGPNIAPSAEE47 pKa = 3.97GLDD50 pKa = 4.01RR51 pKa = 11.84LPSDD55 pKa = 3.46NAQHH59 pKa = 6.72FEE61 pKa = 4.1QAPSPSVQGEE71 pKa = 4.52GVHH74 pKa = 6.16NAPAVGGAAGVGLLGNGAAHH94 pKa = 7.23AGKK97 pKa = 8.58TDD99 pKa = 3.72AEE101 pKa = 4.35CAAAVIGGSSFEE113 pKa = 4.05LRR115 pKa = 11.84FDD117 pKa = 3.83AFVALCSRR125 pKa = 11.84LRR127 pKa = 11.84PEE129 pKa = 4.75PISRR133 pKa = 11.84AVCEE137 pKa = 4.0FRR139 pKa = 11.84LLNGGLNLDD148 pKa = 3.99EE149 pKa = 4.36RR150 pKa = 11.84VAMARR155 pKa = 11.84CVEE158 pKa = 4.18YY159 pKa = 10.77VDD161 pKa = 4.53QYY163 pKa = 11.11EE164 pKa = 4.44FRR166 pKa = 11.84CPTIAVGALTWDD178 pKa = 3.89DD179 pKa = 4.02LVEE182 pKa = 4.28EE183 pKa = 4.34LASTVRR189 pKa = 11.84ADD191 pKa = 4.47PIEE194 pKa = 4.68GPLNQLRR201 pKa = 11.84PVRR204 pKa = 11.84PRR206 pKa = 11.84PSSPPTAVAAAIPVAVPAPEE226 pKa = 4.31LRR228 pKa = 11.84PISWAAASGRR238 pKa = 11.84AEE240 pKa = 3.95PKK242 pKa = 9.94QLQFAADD249 pKa = 4.24DD250 pKa = 4.12DD251 pKa = 4.54CVPDD255 pKa = 3.71VAHH258 pKa = 7.56DD259 pKa = 3.41SGIRR263 pKa = 11.84VGFVDD268 pKa = 4.92EE269 pKa = 4.45PQVAYY274 pKa = 10.44RR275 pKa = 11.84PADD278 pKa = 3.22SDD280 pKa = 3.99YY281 pKa = 11.55DD282 pKa = 4.19DD283 pKa = 4.98EE284 pKa = 5.32PEE286 pKa = 4.21PSNPIVAPSKK296 pKa = 10.28PQMLSQQRR304 pKa = 11.84GFFWRR309 pKa = 11.84MLDD312 pKa = 3.36SVLPRR317 pKa = 11.84FTIFRR322 pKa = 11.84QAATQLHH329 pKa = 6.11ADD331 pKa = 3.73EE332 pKa = 5.04VKK334 pKa = 10.69VSFRR338 pKa = 11.84NASVDD343 pKa = 3.67DD344 pKa = 4.21EE345 pKa = 4.82PEE347 pKa = 4.59LGEE350 pKa = 4.29VAQHH354 pKa = 6.79RR355 pKa = 11.84FRR357 pKa = 11.84DD358 pKa = 3.78VALGAVSHH366 pKa = 6.58RR367 pKa = 11.84KK368 pKa = 6.84YY369 pKa = 10.56HH370 pKa = 6.02RR371 pKa = 11.84AANRR375 pKa = 11.84WHH377 pKa = 5.46QRR379 pKa = 11.84LRR381 pKa = 11.84LLRR384 pKa = 11.84QLRR387 pKa = 11.84EE388 pKa = 3.76EE389 pKa = 5.06LIFEE393 pKa = 4.07SSEE396 pKa = 4.02VKK398 pKa = 10.08RR399 pKa = 11.84VRR401 pKa = 11.84TDD403 pKa = 3.96ANVAWIAIGARR414 pKa = 11.84KK415 pKa = 9.1LVAKK419 pKa = 9.96QVEE422 pKa = 4.17EE423 pKa = 4.18GKK425 pKa = 9.92IDD427 pKa = 3.46RR428 pKa = 11.84RR429 pKa = 11.84HH430 pKa = 5.57ARR432 pKa = 11.84WFRR435 pKa = 11.84SALTEE440 pKa = 4.17TFFLKK445 pKa = 10.67DD446 pKa = 3.72DD447 pKa = 4.51DD448 pKa = 5.47DD449 pKa = 4.78EE450 pKa = 5.22FVDD453 pKa = 3.65TLRR456 pKa = 11.84GSFARR461 pKa = 11.84AWW463 pKa = 3.33
MM1 pKa = 7.35SFAYY5 pKa = 9.28TYY7 pKa = 9.72FFQPKK12 pKa = 9.23RR13 pKa = 11.84APTFDD18 pKa = 3.64FSTTRR23 pKa = 11.84RR24 pKa = 11.84LDD26 pKa = 3.36AFNPATALTGGPGPNIAPSAEE47 pKa = 3.97GLDD50 pKa = 4.01RR51 pKa = 11.84LPSDD55 pKa = 3.46NAQHH59 pKa = 6.72FEE61 pKa = 4.1QAPSPSVQGEE71 pKa = 4.52GVHH74 pKa = 6.16NAPAVGGAAGVGLLGNGAAHH94 pKa = 7.23AGKK97 pKa = 8.58TDD99 pKa = 3.72AEE101 pKa = 4.35CAAAVIGGSSFEE113 pKa = 4.05LRR115 pKa = 11.84FDD117 pKa = 3.83AFVALCSRR125 pKa = 11.84LRR127 pKa = 11.84PEE129 pKa = 4.75PISRR133 pKa = 11.84AVCEE137 pKa = 4.0FRR139 pKa = 11.84LLNGGLNLDD148 pKa = 3.99EE149 pKa = 4.36RR150 pKa = 11.84VAMARR155 pKa = 11.84CVEE158 pKa = 4.18YY159 pKa = 10.77VDD161 pKa = 4.53QYY163 pKa = 11.11EE164 pKa = 4.44FRR166 pKa = 11.84CPTIAVGALTWDD178 pKa = 3.89DD179 pKa = 4.02LVEE182 pKa = 4.28EE183 pKa = 4.34LASTVRR189 pKa = 11.84ADD191 pKa = 4.47PIEE194 pKa = 4.68GPLNQLRR201 pKa = 11.84PVRR204 pKa = 11.84PRR206 pKa = 11.84PSSPPTAVAAAIPVAVPAPEE226 pKa = 4.31LRR228 pKa = 11.84PISWAAASGRR238 pKa = 11.84AEE240 pKa = 3.95PKK242 pKa = 9.94QLQFAADD249 pKa = 4.24DD250 pKa = 4.12DD251 pKa = 4.54CVPDD255 pKa = 3.71VAHH258 pKa = 7.56DD259 pKa = 3.41SGIRR263 pKa = 11.84VGFVDD268 pKa = 4.92EE269 pKa = 4.45PQVAYY274 pKa = 10.44RR275 pKa = 11.84PADD278 pKa = 3.22SDD280 pKa = 3.99YY281 pKa = 11.55DD282 pKa = 4.19DD283 pKa = 4.98EE284 pKa = 5.32PEE286 pKa = 4.21PSNPIVAPSKK296 pKa = 10.28PQMLSQQRR304 pKa = 11.84GFFWRR309 pKa = 11.84MLDD312 pKa = 3.36SVLPRR317 pKa = 11.84FTIFRR322 pKa = 11.84QAATQLHH329 pKa = 6.11ADD331 pKa = 3.73EE332 pKa = 5.04VKK334 pKa = 10.69VSFRR338 pKa = 11.84NASVDD343 pKa = 3.67DD344 pKa = 4.21EE345 pKa = 4.82PEE347 pKa = 4.59LGEE350 pKa = 4.29VAQHH354 pKa = 6.79RR355 pKa = 11.84FRR357 pKa = 11.84DD358 pKa = 3.78VALGAVSHH366 pKa = 6.58RR367 pKa = 11.84KK368 pKa = 6.84YY369 pKa = 10.56HH370 pKa = 6.02RR371 pKa = 11.84AANRR375 pKa = 11.84WHH377 pKa = 5.46QRR379 pKa = 11.84LRR381 pKa = 11.84LLRR384 pKa = 11.84QLRR387 pKa = 11.84EE388 pKa = 3.76EE389 pKa = 5.06LIFEE393 pKa = 4.07SSEE396 pKa = 4.02VKK398 pKa = 10.08RR399 pKa = 11.84VRR401 pKa = 11.84TDD403 pKa = 3.96ANVAWIAIGARR414 pKa = 11.84KK415 pKa = 9.1LVAKK419 pKa = 9.96QVEE422 pKa = 4.17EE423 pKa = 4.18GKK425 pKa = 9.92IDD427 pKa = 3.46RR428 pKa = 11.84RR429 pKa = 11.84HH430 pKa = 5.57ARR432 pKa = 11.84WFRR435 pKa = 11.84SALTEE440 pKa = 4.17TFFLKK445 pKa = 10.67DD446 pKa = 3.72DD447 pKa = 4.51DD448 pKa = 5.47DD449 pKa = 4.78EE450 pKa = 5.22FVDD453 pKa = 3.65TLRR456 pKa = 11.84GSFARR461 pKa = 11.84AWW463 pKa = 3.33
Molecular weight: 51.02 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGI0|A0A1L3KGI0_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 7 OX=1923294 PE=4 SV=1
MM1 pKa = 7.78PMGGLWSATPVLPSVRR17 pKa = 11.84TPWRR21 pKa = 11.84WEE23 pKa = 3.66TLLNCTSSGFTASVLLGCHH42 pKa = 4.74STGTCRR48 pKa = 11.84CFVLCIPACEE58 pKa = 4.36SVGSRR63 pKa = 11.84VGLASPIRR71 pKa = 11.84LGTPVLCNSRR81 pKa = 11.84YY82 pKa = 9.89ARR84 pKa = 11.84QALLLSCLPDD94 pKa = 3.89LLLTNAAYY102 pKa = 10.01HH103 pKa = 6.3LPRR106 pKa = 11.84RR107 pKa = 11.84SISAPHH113 pKa = 6.47CRR115 pKa = 11.84PFWRR119 pKa = 11.84PSSFVLTLTVSLMQTSAQSSRR140 pKa = 11.84FWPDD144 pKa = 2.62HH145 pKa = 6.4RR146 pKa = 11.84GMPRR150 pKa = 11.84NAKK153 pKa = 7.91TANPPASKK161 pKa = 10.34GGQSGRR167 pKa = 11.84QTTPSSRR174 pKa = 11.84PRR176 pKa = 11.84GGTAQKK182 pKa = 10.46VRR184 pKa = 11.84GKK186 pKa = 10.93GEE188 pKa = 3.88YY189 pKa = 10.3LLPGTFAHH197 pKa = 6.8VGRR200 pKa = 11.84KK201 pKa = 9.44LGEE204 pKa = 4.12KK205 pKa = 10.3AGGWLGGGAGALLSTVTGMGDD226 pKa = 3.35YY227 pKa = 10.94QMNNIMHH234 pKa = 7.91ADD236 pKa = 3.86RR237 pKa = 11.84ALTSAHH243 pKa = 6.01PKK245 pKa = 10.26SYY247 pKa = 10.8EE248 pKa = 3.83FTNTEE253 pKa = 3.88LVQVVASTGEE263 pKa = 3.93AFQSYY268 pKa = 8.91QYY270 pKa = 11.51ANNPGLGNFPWLSAIAQRR288 pKa = 11.84FNKK291 pKa = 9.79YY292 pKa = 8.84RR293 pKa = 11.84FKK295 pKa = 10.72QLVYY299 pKa = 9.88HH300 pKa = 6.38FEE302 pKa = 4.45STSSEE307 pKa = 4.2YY308 pKa = 10.95AAGAGLGTVAIATNYY323 pKa = 10.17DD324 pKa = 3.14AVDD327 pKa = 3.49RR328 pKa = 11.84EE329 pKa = 4.23FASMVEE335 pKa = 4.22MEE337 pKa = 4.17ATEE340 pKa = 4.07NAVSGKK346 pKa = 9.78PSVDD350 pKa = 2.92KK351 pKa = 11.37LHH353 pKa = 6.66GVEE356 pKa = 5.16CATAEE361 pKa = 4.19NALKK365 pKa = 8.99WWYY368 pKa = 9.79VRR370 pKa = 11.84SGPPPASTDD379 pKa = 2.8LRR381 pKa = 11.84MYY383 pKa = 11.26DD384 pKa = 3.31MSEE387 pKa = 3.87TTLATEE393 pKa = 4.85GLFASPGTVIGRR405 pKa = 11.84LKK407 pKa = 10.77VFYY410 pKa = 9.73TVEE413 pKa = 4.04FCNPIALGIPAPYY426 pKa = 10.24LPAPVTLRR434 pKa = 11.84SWYY437 pKa = 10.64APGAAAAGVGPYY449 pKa = 10.03FGLKK453 pKa = 10.37DD454 pKa = 4.16SISSPTWSVGPYY466 pKa = 7.74QTLPVNVSGGSVGSIGVTTYY486 pKa = 10.68PVVDD490 pKa = 3.56VTTNTLKK497 pKa = 10.62FYY499 pKa = 11.02CRR501 pKa = 11.84GTFYY505 pKa = 10.14VTARR509 pKa = 11.84FFFSTAPTVTTIAQSTSGACTLNNPYY535 pKa = 10.47SAGNTWHH542 pKa = 6.07VTTSTDD548 pKa = 2.97KK549 pKa = 10.85TLLSVEE555 pKa = 4.14GVITVTSASYY565 pKa = 7.34TTPASVSLSQSGWGSTFVIGSALTSVSYY593 pKa = 10.86VGQPP597 pKa = 2.88
MM1 pKa = 7.78PMGGLWSATPVLPSVRR17 pKa = 11.84TPWRR21 pKa = 11.84WEE23 pKa = 3.66TLLNCTSSGFTASVLLGCHH42 pKa = 4.74STGTCRR48 pKa = 11.84CFVLCIPACEE58 pKa = 4.36SVGSRR63 pKa = 11.84VGLASPIRR71 pKa = 11.84LGTPVLCNSRR81 pKa = 11.84YY82 pKa = 9.89ARR84 pKa = 11.84QALLLSCLPDD94 pKa = 3.89LLLTNAAYY102 pKa = 10.01HH103 pKa = 6.3LPRR106 pKa = 11.84RR107 pKa = 11.84SISAPHH113 pKa = 6.47CRR115 pKa = 11.84PFWRR119 pKa = 11.84PSSFVLTLTVSLMQTSAQSSRR140 pKa = 11.84FWPDD144 pKa = 2.62HH145 pKa = 6.4RR146 pKa = 11.84GMPRR150 pKa = 11.84NAKK153 pKa = 7.91TANPPASKK161 pKa = 10.34GGQSGRR167 pKa = 11.84QTTPSSRR174 pKa = 11.84PRR176 pKa = 11.84GGTAQKK182 pKa = 10.46VRR184 pKa = 11.84GKK186 pKa = 10.93GEE188 pKa = 3.88YY189 pKa = 10.3LLPGTFAHH197 pKa = 6.8VGRR200 pKa = 11.84KK201 pKa = 9.44LGEE204 pKa = 4.12KK205 pKa = 10.3AGGWLGGGAGALLSTVTGMGDD226 pKa = 3.35YY227 pKa = 10.94QMNNIMHH234 pKa = 7.91ADD236 pKa = 3.86RR237 pKa = 11.84ALTSAHH243 pKa = 6.01PKK245 pKa = 10.26SYY247 pKa = 10.8EE248 pKa = 3.83FTNTEE253 pKa = 3.88LVQVVASTGEE263 pKa = 3.93AFQSYY268 pKa = 8.91QYY270 pKa = 11.51ANNPGLGNFPWLSAIAQRR288 pKa = 11.84FNKK291 pKa = 9.79YY292 pKa = 8.84RR293 pKa = 11.84FKK295 pKa = 10.72QLVYY299 pKa = 9.88HH300 pKa = 6.38FEE302 pKa = 4.45STSSEE307 pKa = 4.2YY308 pKa = 10.95AAGAGLGTVAIATNYY323 pKa = 10.17DD324 pKa = 3.14AVDD327 pKa = 3.49RR328 pKa = 11.84EE329 pKa = 4.23FASMVEE335 pKa = 4.22MEE337 pKa = 4.17ATEE340 pKa = 4.07NAVSGKK346 pKa = 9.78PSVDD350 pKa = 2.92KK351 pKa = 11.37LHH353 pKa = 6.66GVEE356 pKa = 5.16CATAEE361 pKa = 4.19NALKK365 pKa = 8.99WWYY368 pKa = 9.79VRR370 pKa = 11.84SGPPPASTDD379 pKa = 2.8LRR381 pKa = 11.84MYY383 pKa = 11.26DD384 pKa = 3.31MSEE387 pKa = 3.87TTLATEE393 pKa = 4.85GLFASPGTVIGRR405 pKa = 11.84LKK407 pKa = 10.77VFYY410 pKa = 9.73TVEE413 pKa = 4.04FCNPIALGIPAPYY426 pKa = 10.24LPAPVTLRR434 pKa = 11.84SWYY437 pKa = 10.64APGAAAAGVGPYY449 pKa = 10.03FGLKK453 pKa = 10.37DD454 pKa = 4.16SISSPTWSVGPYY466 pKa = 7.74QTLPVNVSGGSVGSIGVTTYY486 pKa = 10.68PVVDD490 pKa = 3.56VTTNTLKK497 pKa = 10.62FYY499 pKa = 11.02CRR501 pKa = 11.84GTFYY505 pKa = 10.14VTARR509 pKa = 11.84FFFSTAPTVTTIAQSTSGACTLNNPYY535 pKa = 10.47SAGNTWHH542 pKa = 6.07VTTSTDD548 pKa = 2.97KK549 pKa = 10.85TLLSVEE555 pKa = 4.14GVITVTSASYY565 pKa = 7.34TTPASVSLSQSGWGSTFVIGSALTSVSYY593 pKa = 10.86VGQPP597 pKa = 2.88
Molecular weight: 63.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1501 |
441 |
597 |
500.3 |
54.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.593 ± 1.089 | 1.799 ± 0.211 |
4.863 ± 1.363 | 5.263 ± 0.98 |
4.664 ± 0.427 | 7.662 ± 0.91 |
1.799 ± 0.13 | 2.665 ± 0.165 |
3.131 ± 0.467 | 8.794 ± 0.634 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.865 ± 0.412 | 3.131 ± 0.221 |
6.063 ± 0.96 | 3.464 ± 0.37 |
7.395 ± 1.205 | 7.861 ± 1.27 |
6.262 ± 1.865 | 7.861 ± 0.103 |
1.865 ± 0.162 | 2.998 ± 0.599 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
