
Rodent Torque teno virus 4
Taxonomy: Viruses; Anelloviridae; unclassified Anelloviridae
Average proteome isoelectric point is 8.81
Get precalculated fractions of proteins
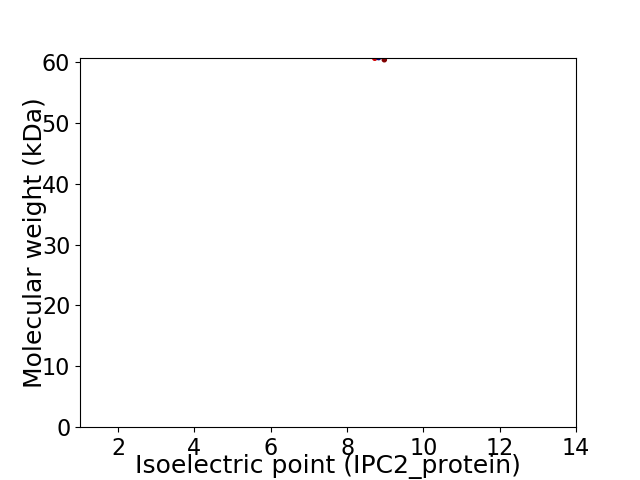
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H4QBT7|A0A2H4QBT7_9VIRU Capsid protein OS=Rodent Torque teno virus 4 OX=2054611 PE=3 SV=1
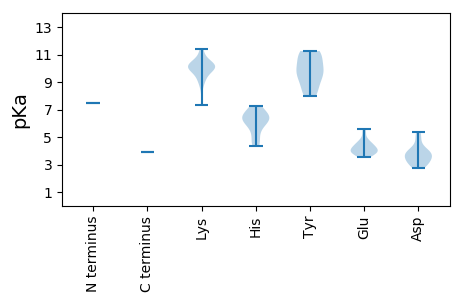
MM1 pKa = 7.49LRR3 pKa = 11.84RR4 pKa = 11.84KK5 pKa = 7.3TRR7 pKa = 11.84HH8 pKa = 4.98VPVMQWNPINRR19 pKa = 11.84KK20 pKa = 8.52KK21 pKa = 9.96CTIRR25 pKa = 11.84GFCPLLYY32 pKa = 10.61CLGGQKK38 pKa = 10.87GIMDD42 pKa = 4.02FTYY45 pKa = 9.9PANKK49 pKa = 10.17AILNWMGGGVHH60 pKa = 6.64SSLISLLDD68 pKa = 3.75LFWEE72 pKa = 4.1EE73 pKa = 4.41RR74 pKa = 11.84YY75 pKa = 8.89WRR77 pKa = 11.84ARR79 pKa = 11.84WSSSNQGFNLFRR91 pKa = 11.84YY92 pKa = 9.11YY93 pKa = 11.13GVTITLYY100 pKa = 8.28PCMYY104 pKa = 7.98YY105 pKa = 10.03TYY107 pKa = 9.79IFWYY111 pKa = 8.27STEE114 pKa = 3.88EE115 pKa = 3.83LAEE118 pKa = 4.51DD119 pKa = 4.07SEE121 pKa = 4.75PLTVCHH127 pKa = 6.82PSQLLLSKK135 pKa = 10.23HH136 pKa = 5.56HH137 pKa = 6.36VIVHH141 pKa = 6.44RR142 pKa = 11.84MHH144 pKa = 6.2GMKK147 pKa = 10.19VKK149 pKa = 9.95PVKK152 pKa = 10.46LRR154 pKa = 11.84IKK156 pKa = 10.04PPSQMTGTWHH166 pKa = 7.26SFAKK170 pKa = 9.25WAKK173 pKa = 9.75QPLLKK178 pKa = 10.09WRR180 pKa = 11.84VSLCDD185 pKa = 4.57IEE187 pKa = 5.3NPWTGFPNSEE197 pKa = 3.98TQGIHH202 pKa = 4.57VKK204 pKa = 9.7VWAISTTSTAAGEE217 pKa = 3.75KK218 pKa = 10.13DD219 pKa = 2.73IWYY222 pKa = 9.35FPLLDD227 pKa = 5.36DD228 pKa = 5.03GTDD231 pKa = 2.96VWICLHH237 pKa = 5.8EE238 pKa = 4.59IKK240 pKa = 10.06WNRR243 pKa = 11.84NGDD246 pKa = 3.84GPDD249 pKa = 3.32EE250 pKa = 4.62SSSNFWPVQAEE261 pKa = 4.2FQKK264 pKa = 11.24LLVPFYY270 pKa = 9.58MYY272 pKa = 11.21GFGRR276 pKa = 11.84SATFYY281 pKa = 11.25LMQKK285 pKa = 9.04DD286 pKa = 3.86VHH288 pKa = 6.22MPAPSQGDD296 pKa = 3.01KK297 pKa = 11.42GKK299 pKa = 10.3FLFVKK304 pKa = 9.18TINSPAWRR312 pKa = 11.84GADD315 pKa = 3.09GGFPQFKK322 pKa = 10.52DD323 pKa = 3.08NQFYY327 pKa = 10.69MKK329 pKa = 9.9FSTVAQIAAQGPWCMKK345 pKa = 10.27SIGNEE350 pKa = 3.84THH352 pKa = 6.5GVNLSMKK359 pKa = 10.25YY360 pKa = 9.85KK361 pKa = 10.62FHH363 pKa = 6.2FQWGGTPGTHH373 pKa = 7.08LPPVQPADD381 pKa = 3.75GGPPQPLLQSTIRR394 pKa = 11.84WGNSLRR400 pKa = 11.84ADD402 pKa = 3.61LRR404 pKa = 11.84DD405 pKa = 3.53PTTVGDD411 pKa = 4.0EE412 pKa = 4.19VLHH415 pKa = 6.38PEE417 pKa = 4.33EE418 pKa = 5.55LDD420 pKa = 3.39SAGIINPRR428 pKa = 11.84AFRR431 pKa = 11.84RR432 pKa = 11.84ITKK435 pKa = 9.2PAVSTGSRR443 pKa = 11.84SVGTLGCLRR452 pKa = 11.84PAVWSAEE459 pKa = 3.74KK460 pKa = 10.24RR461 pKa = 11.84QEE463 pKa = 3.99PSSEE467 pKa = 3.54EE468 pKa = 4.14SAQEE472 pKa = 4.3EE473 pKa = 4.87YY474 pKa = 10.96SSSEE478 pKa = 4.08TGSEE482 pKa = 4.4TEE484 pKa = 3.97TEE486 pKa = 3.94EE487 pKa = 3.71MDD489 pKa = 3.44RR490 pKa = 11.84RR491 pKa = 11.84RR492 pKa = 11.84HH493 pKa = 4.36QRR495 pKa = 11.84RR496 pKa = 11.84KK497 pKa = 9.91RR498 pKa = 11.84KK499 pKa = 9.71RR500 pKa = 11.84LEE502 pKa = 3.52QLLRR506 pKa = 11.84GLMTVGLKK514 pKa = 10.22RR515 pKa = 11.84PMMAGPSGSSQRR527 pKa = 11.84SVSII531 pKa = 3.88
MM1 pKa = 7.49LRR3 pKa = 11.84RR4 pKa = 11.84KK5 pKa = 7.3TRR7 pKa = 11.84HH8 pKa = 4.98VPVMQWNPINRR19 pKa = 11.84KK20 pKa = 8.52KK21 pKa = 9.96CTIRR25 pKa = 11.84GFCPLLYY32 pKa = 10.61CLGGQKK38 pKa = 10.87GIMDD42 pKa = 4.02FTYY45 pKa = 9.9PANKK49 pKa = 10.17AILNWMGGGVHH60 pKa = 6.64SSLISLLDD68 pKa = 3.75LFWEE72 pKa = 4.1EE73 pKa = 4.41RR74 pKa = 11.84YY75 pKa = 8.89WRR77 pKa = 11.84ARR79 pKa = 11.84WSSSNQGFNLFRR91 pKa = 11.84YY92 pKa = 9.11YY93 pKa = 11.13GVTITLYY100 pKa = 8.28PCMYY104 pKa = 7.98YY105 pKa = 10.03TYY107 pKa = 9.79IFWYY111 pKa = 8.27STEE114 pKa = 3.88EE115 pKa = 3.83LAEE118 pKa = 4.51DD119 pKa = 4.07SEE121 pKa = 4.75PLTVCHH127 pKa = 6.82PSQLLLSKK135 pKa = 10.23HH136 pKa = 5.56HH137 pKa = 6.36VIVHH141 pKa = 6.44RR142 pKa = 11.84MHH144 pKa = 6.2GMKK147 pKa = 10.19VKK149 pKa = 9.95PVKK152 pKa = 10.46LRR154 pKa = 11.84IKK156 pKa = 10.04PPSQMTGTWHH166 pKa = 7.26SFAKK170 pKa = 9.25WAKK173 pKa = 9.75QPLLKK178 pKa = 10.09WRR180 pKa = 11.84VSLCDD185 pKa = 4.57IEE187 pKa = 5.3NPWTGFPNSEE197 pKa = 3.98TQGIHH202 pKa = 4.57VKK204 pKa = 9.7VWAISTTSTAAGEE217 pKa = 3.75KK218 pKa = 10.13DD219 pKa = 2.73IWYY222 pKa = 9.35FPLLDD227 pKa = 5.36DD228 pKa = 5.03GTDD231 pKa = 2.96VWICLHH237 pKa = 5.8EE238 pKa = 4.59IKK240 pKa = 10.06WNRR243 pKa = 11.84NGDD246 pKa = 3.84GPDD249 pKa = 3.32EE250 pKa = 4.62SSSNFWPVQAEE261 pKa = 4.2FQKK264 pKa = 11.24LLVPFYY270 pKa = 9.58MYY272 pKa = 11.21GFGRR276 pKa = 11.84SATFYY281 pKa = 11.25LMQKK285 pKa = 9.04DD286 pKa = 3.86VHH288 pKa = 6.22MPAPSQGDD296 pKa = 3.01KK297 pKa = 11.42GKK299 pKa = 10.3FLFVKK304 pKa = 9.18TINSPAWRR312 pKa = 11.84GADD315 pKa = 3.09GGFPQFKK322 pKa = 10.52DD323 pKa = 3.08NQFYY327 pKa = 10.69MKK329 pKa = 9.9FSTVAQIAAQGPWCMKK345 pKa = 10.27SIGNEE350 pKa = 3.84THH352 pKa = 6.5GVNLSMKK359 pKa = 10.25YY360 pKa = 9.85KK361 pKa = 10.62FHH363 pKa = 6.2FQWGGTPGTHH373 pKa = 7.08LPPVQPADD381 pKa = 3.75GGPPQPLLQSTIRR394 pKa = 11.84WGNSLRR400 pKa = 11.84ADD402 pKa = 3.61LRR404 pKa = 11.84DD405 pKa = 3.53PTTVGDD411 pKa = 4.0EE412 pKa = 4.19VLHH415 pKa = 6.38PEE417 pKa = 4.33EE418 pKa = 5.55LDD420 pKa = 3.39SAGIINPRR428 pKa = 11.84AFRR431 pKa = 11.84RR432 pKa = 11.84ITKK435 pKa = 9.2PAVSTGSRR443 pKa = 11.84SVGTLGCLRR452 pKa = 11.84PAVWSAEE459 pKa = 3.74KK460 pKa = 10.24RR461 pKa = 11.84QEE463 pKa = 3.99PSSEE467 pKa = 3.54EE468 pKa = 4.14SAQEE472 pKa = 4.3EE473 pKa = 4.87YY474 pKa = 10.96SSSEE478 pKa = 4.08TGSEE482 pKa = 4.4TEE484 pKa = 3.97TEE486 pKa = 3.94EE487 pKa = 3.71MDD489 pKa = 3.44RR490 pKa = 11.84RR491 pKa = 11.84RR492 pKa = 11.84HH493 pKa = 4.36QRR495 pKa = 11.84RR496 pKa = 11.84KK497 pKa = 9.91RR498 pKa = 11.84KK499 pKa = 9.71RR500 pKa = 11.84LEE502 pKa = 3.52QLLRR506 pKa = 11.84GLMTVGLKK514 pKa = 10.22RR515 pKa = 11.84PMMAGPSGSSQRR527 pKa = 11.84SVSII531 pKa = 3.88
Molecular weight: 60.59 kDa
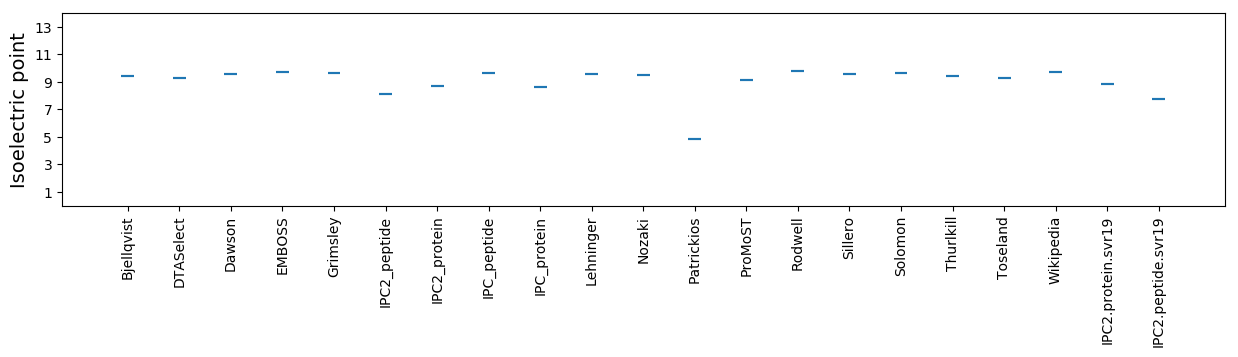
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H4QBT7|A0A2H4QBT7_9VIRU Capsid protein OS=Rodent Torque teno virus 4 OX=2054611 PE=3 SV=1
MM1 pKa = 7.49LRR3 pKa = 11.84RR4 pKa = 11.84KK5 pKa = 7.3TRR7 pKa = 11.84HH8 pKa = 4.98VPVMQWNPINRR19 pKa = 11.84KK20 pKa = 8.52KK21 pKa = 9.96CTIRR25 pKa = 11.84GFCPLLYY32 pKa = 10.61CLGGQKK38 pKa = 10.87GIMDD42 pKa = 4.02FTYY45 pKa = 9.9PANKK49 pKa = 10.17AILNWMGGGVHH60 pKa = 6.64SSLISLLDD68 pKa = 3.75LFWEE72 pKa = 4.1EE73 pKa = 4.41RR74 pKa = 11.84YY75 pKa = 8.89WRR77 pKa = 11.84ARR79 pKa = 11.84WSSSNQGFNLFRR91 pKa = 11.84YY92 pKa = 9.11YY93 pKa = 11.13GVTITLYY100 pKa = 8.28PCMYY104 pKa = 7.98YY105 pKa = 10.03TYY107 pKa = 9.79IFWYY111 pKa = 8.27STEE114 pKa = 3.88EE115 pKa = 3.83LAEE118 pKa = 4.51DD119 pKa = 4.07SEE121 pKa = 4.75PLTVCHH127 pKa = 6.82PSQLLLSKK135 pKa = 10.23HH136 pKa = 5.56HH137 pKa = 6.36VIVHH141 pKa = 6.44RR142 pKa = 11.84MHH144 pKa = 6.2GMKK147 pKa = 10.19VKK149 pKa = 9.95PVKK152 pKa = 10.46LRR154 pKa = 11.84IKK156 pKa = 10.04PPSQMTGTWHH166 pKa = 7.26SFAKK170 pKa = 9.25WAKK173 pKa = 9.75QPLLKK178 pKa = 10.09WRR180 pKa = 11.84VSLCDD185 pKa = 4.57IEE187 pKa = 5.3NPWTGFPNSEE197 pKa = 3.98TQGIHH202 pKa = 4.57VKK204 pKa = 9.7VWAISTTSTAAGEE217 pKa = 3.75KK218 pKa = 10.13DD219 pKa = 2.73IWYY222 pKa = 9.35FPLLDD227 pKa = 5.36DD228 pKa = 5.03GTDD231 pKa = 2.96VWICLHH237 pKa = 5.8EE238 pKa = 4.59IKK240 pKa = 10.06WNRR243 pKa = 11.84NGDD246 pKa = 3.84GPDD249 pKa = 3.32EE250 pKa = 4.62SSSNFWPVQAEE261 pKa = 4.2FQKK264 pKa = 11.24LLVPFYY270 pKa = 9.58MYY272 pKa = 11.21GFGRR276 pKa = 11.84SATFYY281 pKa = 11.25LMQKK285 pKa = 9.04DD286 pKa = 3.86VHH288 pKa = 6.22MPAPSQGDD296 pKa = 3.01KK297 pKa = 11.42GKK299 pKa = 10.3FLFVKK304 pKa = 9.18TINSPAWRR312 pKa = 11.84GADD315 pKa = 3.09GGFPQFKK322 pKa = 10.52DD323 pKa = 3.08NQFYY327 pKa = 10.69MKK329 pKa = 9.9FSTVAQIAAQGPWCMKK345 pKa = 10.27SIGNEE350 pKa = 3.84THH352 pKa = 6.5GVNLSMKK359 pKa = 10.25YY360 pKa = 9.85KK361 pKa = 10.62FHH363 pKa = 6.2FQWGGTPGTHH373 pKa = 7.08LPPVQPADD381 pKa = 3.75GGPPQPLLQSTIRR394 pKa = 11.84WGNSLRR400 pKa = 11.84ADD402 pKa = 3.61LRR404 pKa = 11.84DD405 pKa = 3.53PTTVGDD411 pKa = 4.0EE412 pKa = 4.19VLHH415 pKa = 6.38PEE417 pKa = 4.33EE418 pKa = 5.55LDD420 pKa = 3.39SAGIINPRR428 pKa = 11.84AFRR431 pKa = 11.84RR432 pKa = 11.84ITKK435 pKa = 9.2PAVSTGSRR443 pKa = 11.84SVGTLGCLRR452 pKa = 11.84PAVWSAEE459 pKa = 3.74KK460 pKa = 10.24RR461 pKa = 11.84QEE463 pKa = 3.99PSSEE467 pKa = 3.54EE468 pKa = 4.14SAQEE472 pKa = 4.3EE473 pKa = 4.87YY474 pKa = 10.96SSSEE478 pKa = 4.08TGSEE482 pKa = 4.4TEE484 pKa = 3.97TEE486 pKa = 3.94EE487 pKa = 3.71MDD489 pKa = 3.44RR490 pKa = 11.84RR491 pKa = 11.84RR492 pKa = 11.84HH493 pKa = 4.36QRR495 pKa = 11.84RR496 pKa = 11.84KK497 pKa = 9.91RR498 pKa = 11.84KK499 pKa = 9.71RR500 pKa = 11.84LEE502 pKa = 3.52QLLRR506 pKa = 11.84GLMTVGLKK514 pKa = 10.22RR515 pKa = 11.84PMMAGPSGSSQRR527 pKa = 11.84SVSII531 pKa = 3.88
MM1 pKa = 7.49LRR3 pKa = 11.84RR4 pKa = 11.84KK5 pKa = 7.3TRR7 pKa = 11.84HH8 pKa = 4.98VPVMQWNPINRR19 pKa = 11.84KK20 pKa = 8.52KK21 pKa = 9.96CTIRR25 pKa = 11.84GFCPLLYY32 pKa = 10.61CLGGQKK38 pKa = 10.87GIMDD42 pKa = 4.02FTYY45 pKa = 9.9PANKK49 pKa = 10.17AILNWMGGGVHH60 pKa = 6.64SSLISLLDD68 pKa = 3.75LFWEE72 pKa = 4.1EE73 pKa = 4.41RR74 pKa = 11.84YY75 pKa = 8.89WRR77 pKa = 11.84ARR79 pKa = 11.84WSSSNQGFNLFRR91 pKa = 11.84YY92 pKa = 9.11YY93 pKa = 11.13GVTITLYY100 pKa = 8.28PCMYY104 pKa = 7.98YY105 pKa = 10.03TYY107 pKa = 9.79IFWYY111 pKa = 8.27STEE114 pKa = 3.88EE115 pKa = 3.83LAEE118 pKa = 4.51DD119 pKa = 4.07SEE121 pKa = 4.75PLTVCHH127 pKa = 6.82PSQLLLSKK135 pKa = 10.23HH136 pKa = 5.56HH137 pKa = 6.36VIVHH141 pKa = 6.44RR142 pKa = 11.84MHH144 pKa = 6.2GMKK147 pKa = 10.19VKK149 pKa = 9.95PVKK152 pKa = 10.46LRR154 pKa = 11.84IKK156 pKa = 10.04PPSQMTGTWHH166 pKa = 7.26SFAKK170 pKa = 9.25WAKK173 pKa = 9.75QPLLKK178 pKa = 10.09WRR180 pKa = 11.84VSLCDD185 pKa = 4.57IEE187 pKa = 5.3NPWTGFPNSEE197 pKa = 3.98TQGIHH202 pKa = 4.57VKK204 pKa = 9.7VWAISTTSTAAGEE217 pKa = 3.75KK218 pKa = 10.13DD219 pKa = 2.73IWYY222 pKa = 9.35FPLLDD227 pKa = 5.36DD228 pKa = 5.03GTDD231 pKa = 2.96VWICLHH237 pKa = 5.8EE238 pKa = 4.59IKK240 pKa = 10.06WNRR243 pKa = 11.84NGDD246 pKa = 3.84GPDD249 pKa = 3.32EE250 pKa = 4.62SSSNFWPVQAEE261 pKa = 4.2FQKK264 pKa = 11.24LLVPFYY270 pKa = 9.58MYY272 pKa = 11.21GFGRR276 pKa = 11.84SATFYY281 pKa = 11.25LMQKK285 pKa = 9.04DD286 pKa = 3.86VHH288 pKa = 6.22MPAPSQGDD296 pKa = 3.01KK297 pKa = 11.42GKK299 pKa = 10.3FLFVKK304 pKa = 9.18TINSPAWRR312 pKa = 11.84GADD315 pKa = 3.09GGFPQFKK322 pKa = 10.52DD323 pKa = 3.08NQFYY327 pKa = 10.69MKK329 pKa = 9.9FSTVAQIAAQGPWCMKK345 pKa = 10.27SIGNEE350 pKa = 3.84THH352 pKa = 6.5GVNLSMKK359 pKa = 10.25YY360 pKa = 9.85KK361 pKa = 10.62FHH363 pKa = 6.2FQWGGTPGTHH373 pKa = 7.08LPPVQPADD381 pKa = 3.75GGPPQPLLQSTIRR394 pKa = 11.84WGNSLRR400 pKa = 11.84ADD402 pKa = 3.61LRR404 pKa = 11.84DD405 pKa = 3.53PTTVGDD411 pKa = 4.0EE412 pKa = 4.19VLHH415 pKa = 6.38PEE417 pKa = 4.33EE418 pKa = 5.55LDD420 pKa = 3.39SAGIINPRR428 pKa = 11.84AFRR431 pKa = 11.84RR432 pKa = 11.84ITKK435 pKa = 9.2PAVSTGSRR443 pKa = 11.84SVGTLGCLRR452 pKa = 11.84PAVWSAEE459 pKa = 3.74KK460 pKa = 10.24RR461 pKa = 11.84QEE463 pKa = 3.99PSSEE467 pKa = 3.54EE468 pKa = 4.14SAQEE472 pKa = 4.3EE473 pKa = 4.87YY474 pKa = 10.96SSSEE478 pKa = 4.08TGSEE482 pKa = 4.4TEE484 pKa = 3.97TEE486 pKa = 3.94EE487 pKa = 3.71MDD489 pKa = 3.44RR490 pKa = 11.84RR491 pKa = 11.84RR492 pKa = 11.84HH493 pKa = 4.36QRR495 pKa = 11.84RR496 pKa = 11.84KK497 pKa = 9.91RR498 pKa = 11.84KK499 pKa = 9.71RR500 pKa = 11.84LEE502 pKa = 3.52QLLRR506 pKa = 11.84GLMTVGLKK514 pKa = 10.22RR515 pKa = 11.84PMMAGPSGSSQRR527 pKa = 11.84SVSII531 pKa = 3.88
Molecular weight: 60.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
531 |
531 |
531 |
531.0 |
60.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.896 ± 0.0 | 1.695 ± 0.0 |
3.766 ± 0.0 | 5.273 ± 0.0 |
4.331 ± 0.0 | 8.098 ± 0.0 |
3.013 ± 0.0 | 4.331 ± 0.0 |
5.838 ± 0.0 | 7.91 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.39 ± 0.0 | 3.202 ± 0.0 |
6.968 ± 0.0 | 4.52 ± 0.0 |
6.403 ± 0.0 | 8.286 ± 0.0 |
6.026 ± 0.0 | 5.085 ± 0.0 |
3.766 ± 0.0 | 3.202 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
