
Torque teno indri virus 1
Taxonomy: Viruses; Anelloviridae; Chitorquevirus; Torque teno indriid virus 1
Average proteome isoelectric point is 7.5
Get precalculated fractions of proteins
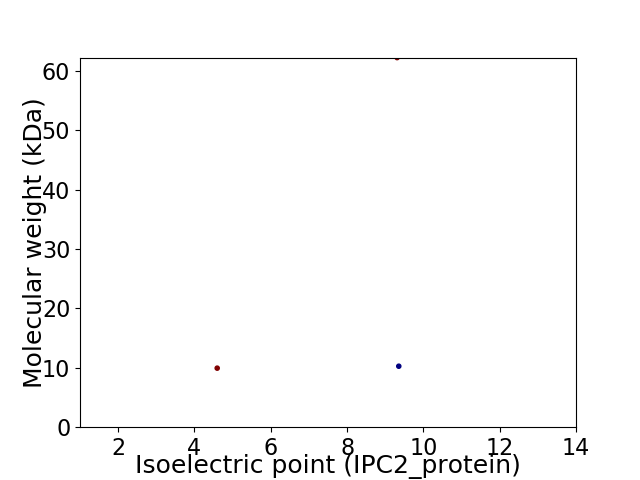
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
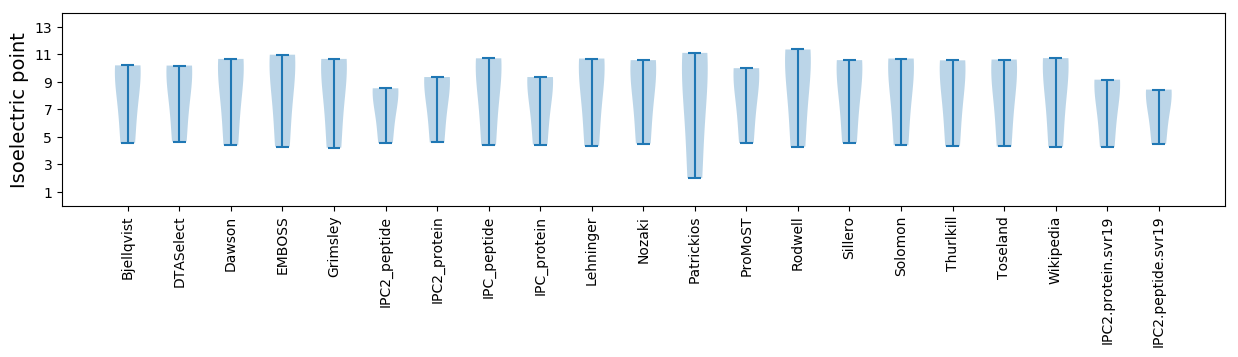
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1Z1W0S6|A0A1Z1W0S6_9VIRU Capsid protein OS=Torque teno indri virus 1 OX=2010248 GN=orf1 PE=3 SV=1
MM1 pKa = 8.18DD2 pKa = 4.52YY3 pKa = 11.01SRR5 pKa = 11.84QKK7 pKa = 10.52QNEE10 pKa = 4.38AKK12 pKa = 9.34WKK14 pKa = 10.32LGINLAHH21 pKa = 6.18STWCSCGDD29 pKa = 3.28WFGHH33 pKa = 5.51LTCIYY38 pKa = 8.83YY39 pKa = 10.55RR40 pKa = 11.84SGEE43 pKa = 4.15SSTAVQAKK51 pKa = 9.67NKK53 pKa = 9.27TPEE56 pKa = 3.91TCLGGGGEE64 pKa = 4.32GGVYY68 pKa = 9.81QGGGAGGGEE77 pKa = 4.58DD78 pKa = 3.63GGEE81 pKa = 4.03DD82 pKa = 3.45ATIPDD87 pKa = 4.05AALEE91 pKa = 4.24DD92 pKa = 4.2AEE94 pKa = 4.27QQ95 pKa = 3.66
MM1 pKa = 8.18DD2 pKa = 4.52YY3 pKa = 11.01SRR5 pKa = 11.84QKK7 pKa = 10.52QNEE10 pKa = 4.38AKK12 pKa = 9.34WKK14 pKa = 10.32LGINLAHH21 pKa = 6.18STWCSCGDD29 pKa = 3.28WFGHH33 pKa = 5.51LTCIYY38 pKa = 8.83YY39 pKa = 10.55RR40 pKa = 11.84SGEE43 pKa = 4.15SSTAVQAKK51 pKa = 9.67NKK53 pKa = 9.27TPEE56 pKa = 3.91TCLGGGGEE64 pKa = 4.32GGVYY68 pKa = 9.81QGGGAGGGEE77 pKa = 4.58DD78 pKa = 3.63GGEE81 pKa = 4.03DD82 pKa = 3.45ATIPDD87 pKa = 4.05AALEE91 pKa = 4.24DD92 pKa = 4.2AEE94 pKa = 4.27QQ95 pKa = 3.66
Molecular weight: 9.95 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Z1W0T3|A0A1Z1W0T3_9VIRU ORF3 OS=Torque teno indri virus 1 OX=2010248 GN=orf3 PE=4 SV=1
MM1 pKa = 7.07PWRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84WRR10 pKa = 11.84IPGRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84WRR18 pKa = 11.84WRR20 pKa = 11.84RR21 pKa = 11.84WWRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84YY27 pKa = 8.75YY28 pKa = 9.2PRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84FRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84TVRR40 pKa = 11.84KK41 pKa = 8.16PLNIIQYY48 pKa = 7.23QPRR51 pKa = 11.84IIKK54 pKa = 10.2RR55 pKa = 11.84CVIKK59 pKa = 10.93GKK61 pKa = 10.14VLVLGGHH68 pKa = 6.04TFQTIEE74 pKa = 4.3RR75 pKa = 11.84FHH77 pKa = 6.08HH78 pKa = 6.04TKK80 pKa = 9.25TAADD84 pKa = 3.9GTTEE88 pKa = 4.05TVFVHH93 pKa = 6.94GGTTDD98 pKa = 2.59WMTVTLQEE106 pKa = 4.94LYY108 pKa = 10.64KK109 pKa = 10.67GWKK112 pKa = 8.38QRR114 pKa = 11.84QNTWSTSNATFDD126 pKa = 3.18MARR129 pKa = 11.84YY130 pKa = 9.23KK131 pKa = 10.53GAKK134 pKa = 6.84FTLWKK139 pKa = 10.24HH140 pKa = 5.53IEE142 pKa = 3.82HH143 pKa = 7.53DD144 pKa = 4.39YY145 pKa = 10.39VVVWDD150 pKa = 4.47TEE152 pKa = 4.14QSALLTADD160 pKa = 3.6IQRR163 pKa = 11.84MQPFQAQLTEE173 pKa = 4.09KK174 pKa = 9.7NHH176 pKa = 5.22IVVRR180 pKa = 11.84SNRR183 pKa = 11.84WGNSAPAKK191 pKa = 8.12TLRR194 pKa = 11.84IKK196 pKa = 10.34PPPGMKK202 pKa = 9.8NQWYY206 pKa = 9.15YY207 pKa = 10.8MSDD210 pKa = 3.1IADD213 pKa = 3.68KK214 pKa = 10.71TLVNYY219 pKa = 8.88GFSIADD225 pKa = 3.25IRR227 pKa = 11.84DD228 pKa = 3.26GGAGDD233 pKa = 3.7TGRR236 pKa = 11.84IITPWPDD243 pKa = 2.7GSVEE247 pKa = 4.08NTVTEE252 pKa = 4.21GGKK255 pKa = 10.25KK256 pKa = 7.94YY257 pKa = 9.34TIQNYY262 pKa = 7.37SWQWDD267 pKa = 3.79LPPWQIQISDD277 pKa = 3.3KK278 pKa = 9.99WRR280 pKa = 11.84QPFWMSQRR288 pKa = 11.84GLGLTEE294 pKa = 4.33NNWIYY299 pKa = 10.56DD300 pKa = 3.4INIPYY305 pKa = 8.87YY306 pKa = 8.9QFWWGIQGSEE316 pKa = 4.23DD317 pKa = 3.47PQINTDD323 pKa = 3.16YY324 pKa = 10.32WGKK327 pKa = 10.7KK328 pKa = 7.65EE329 pKa = 5.25DD330 pKa = 3.87GMEE333 pKa = 3.74NLYY336 pKa = 10.03PYY338 pKa = 10.25KK339 pKa = 10.27ILWWDD344 pKa = 3.28QEE346 pKa = 4.09ANRR349 pKa = 11.84RR350 pKa = 11.84LFGIKK355 pKa = 9.94FPGVIHH361 pKa = 7.04ILQNGPMVPHH371 pKa = 7.01LKK373 pKa = 10.6NPFSHH378 pKa = 7.02IIDD381 pKa = 3.57YY382 pKa = 10.55KK383 pKa = 10.68FYY385 pKa = 9.9WEE387 pKa = 4.21WGGHH391 pKa = 5.27IPHH394 pKa = 6.59SRR396 pKa = 11.84NIEE399 pKa = 3.77KK400 pKa = 10.54DD401 pKa = 3.14PRR403 pKa = 11.84FQTFKK408 pKa = 11.05GQDD411 pKa = 3.23VQNPRR416 pKa = 11.84KK417 pKa = 9.88AYY419 pKa = 7.4KK420 pKa = 9.96TSLAPWDD427 pKa = 3.81FDD429 pKa = 4.23KK430 pKa = 11.53DD431 pKa = 3.8GLITKK436 pKa = 9.67HH437 pKa = 5.74AFRR440 pKa = 11.84RR441 pKa = 11.84LLSPTEE447 pKa = 4.4DD448 pKa = 3.62EE449 pKa = 4.87SDD451 pKa = 3.6CSLHH455 pKa = 6.21FKK457 pKa = 10.4EE458 pKa = 5.27RR459 pKa = 11.84VPKK462 pKa = 9.99EE463 pKa = 3.97QEE465 pKa = 3.86DD466 pKa = 4.06DD467 pKa = 3.69SSQEE471 pKa = 3.74EE472 pKa = 4.66SSCNGEE478 pKa = 4.04SDD480 pKa = 3.79SDD482 pKa = 3.99SDD484 pKa = 3.78STLQIPIQKK493 pKa = 9.68QIKK496 pKa = 9.43LIQRR500 pKa = 11.84RR501 pKa = 11.84LAMDD505 pKa = 3.4CQFRR509 pKa = 11.84KK510 pKa = 10.01RR511 pKa = 11.84LRR513 pKa = 11.84TYY515 pKa = 11.18LKK517 pKa = 10.21AQKK520 pKa = 10.39
MM1 pKa = 7.07PWRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84WRR10 pKa = 11.84IPGRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84WRR18 pKa = 11.84WRR20 pKa = 11.84RR21 pKa = 11.84WWRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84YY27 pKa = 8.75YY28 pKa = 9.2PRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84FRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84TVRR40 pKa = 11.84KK41 pKa = 8.16PLNIIQYY48 pKa = 7.23QPRR51 pKa = 11.84IIKK54 pKa = 10.2RR55 pKa = 11.84CVIKK59 pKa = 10.93GKK61 pKa = 10.14VLVLGGHH68 pKa = 6.04TFQTIEE74 pKa = 4.3RR75 pKa = 11.84FHH77 pKa = 6.08HH78 pKa = 6.04TKK80 pKa = 9.25TAADD84 pKa = 3.9GTTEE88 pKa = 4.05TVFVHH93 pKa = 6.94GGTTDD98 pKa = 2.59WMTVTLQEE106 pKa = 4.94LYY108 pKa = 10.64KK109 pKa = 10.67GWKK112 pKa = 8.38QRR114 pKa = 11.84QNTWSTSNATFDD126 pKa = 3.18MARR129 pKa = 11.84YY130 pKa = 9.23KK131 pKa = 10.53GAKK134 pKa = 6.84FTLWKK139 pKa = 10.24HH140 pKa = 5.53IEE142 pKa = 3.82HH143 pKa = 7.53DD144 pKa = 4.39YY145 pKa = 10.39VVVWDD150 pKa = 4.47TEE152 pKa = 4.14QSALLTADD160 pKa = 3.6IQRR163 pKa = 11.84MQPFQAQLTEE173 pKa = 4.09KK174 pKa = 9.7NHH176 pKa = 5.22IVVRR180 pKa = 11.84SNRR183 pKa = 11.84WGNSAPAKK191 pKa = 8.12TLRR194 pKa = 11.84IKK196 pKa = 10.34PPPGMKK202 pKa = 9.8NQWYY206 pKa = 9.15YY207 pKa = 10.8MSDD210 pKa = 3.1IADD213 pKa = 3.68KK214 pKa = 10.71TLVNYY219 pKa = 8.88GFSIADD225 pKa = 3.25IRR227 pKa = 11.84DD228 pKa = 3.26GGAGDD233 pKa = 3.7TGRR236 pKa = 11.84IITPWPDD243 pKa = 2.7GSVEE247 pKa = 4.08NTVTEE252 pKa = 4.21GGKK255 pKa = 10.25KK256 pKa = 7.94YY257 pKa = 9.34TIQNYY262 pKa = 7.37SWQWDD267 pKa = 3.79LPPWQIQISDD277 pKa = 3.3KK278 pKa = 9.99WRR280 pKa = 11.84QPFWMSQRR288 pKa = 11.84GLGLTEE294 pKa = 4.33NNWIYY299 pKa = 10.56DD300 pKa = 3.4INIPYY305 pKa = 8.87YY306 pKa = 8.9QFWWGIQGSEE316 pKa = 4.23DD317 pKa = 3.47PQINTDD323 pKa = 3.16YY324 pKa = 10.32WGKK327 pKa = 10.7KK328 pKa = 7.65EE329 pKa = 5.25DD330 pKa = 3.87GMEE333 pKa = 3.74NLYY336 pKa = 10.03PYY338 pKa = 10.25KK339 pKa = 10.27ILWWDD344 pKa = 3.28QEE346 pKa = 4.09ANRR349 pKa = 11.84RR350 pKa = 11.84LFGIKK355 pKa = 9.94FPGVIHH361 pKa = 7.04ILQNGPMVPHH371 pKa = 7.01LKK373 pKa = 10.6NPFSHH378 pKa = 7.02IIDD381 pKa = 3.57YY382 pKa = 10.55KK383 pKa = 10.68FYY385 pKa = 9.9WEE387 pKa = 4.21WGGHH391 pKa = 5.27IPHH394 pKa = 6.59SRR396 pKa = 11.84NIEE399 pKa = 3.77KK400 pKa = 10.54DD401 pKa = 3.14PRR403 pKa = 11.84FQTFKK408 pKa = 11.05GQDD411 pKa = 3.23VQNPRR416 pKa = 11.84KK417 pKa = 9.88AYY419 pKa = 7.4KK420 pKa = 9.96TSLAPWDD427 pKa = 3.81FDD429 pKa = 4.23KK430 pKa = 11.53DD431 pKa = 3.8GLITKK436 pKa = 9.67HH437 pKa = 5.74AFRR440 pKa = 11.84RR441 pKa = 11.84LLSPTEE447 pKa = 4.4DD448 pKa = 3.62EE449 pKa = 4.87SDD451 pKa = 3.6CSLHH455 pKa = 6.21FKK457 pKa = 10.4EE458 pKa = 5.27RR459 pKa = 11.84VPKK462 pKa = 9.99EE463 pKa = 3.97QEE465 pKa = 3.86DD466 pKa = 4.06DD467 pKa = 3.69SSQEE471 pKa = 3.74EE472 pKa = 4.66SSCNGEE478 pKa = 4.04SDD480 pKa = 3.79SDD482 pKa = 3.99SDD484 pKa = 3.78STLQIPIQKK493 pKa = 9.68QIKK496 pKa = 9.43LIQRR500 pKa = 11.84RR501 pKa = 11.84LAMDD505 pKa = 3.4CQFRR509 pKa = 11.84KK510 pKa = 10.01RR511 pKa = 11.84LRR513 pKa = 11.84TYY515 pKa = 11.18LKK517 pKa = 10.21AQKK520 pKa = 10.39
Molecular weight: 62.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
705 |
90 |
520 |
235.0 |
27.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.965 ± 1.871 | 1.277 ± 0.848 |
6.099 ± 0.526 | 5.106 ± 1.04 |
3.546 ± 0.689 | 7.234 ± 3.434 |
2.553 ± 0.199 | 6.099 ± 1.464 |
7.801 ± 2.046 | 5.39 ± 0.327 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.844 ± 0.222 | 3.83 ± 0.298 |
5.106 ± 0.845 | 6.241 ± 0.425 |
8.085 ± 1.979 | 6.95 ± 3.869 |
6.667 ± 0.343 | 2.979 ± 1.099 |
4.681 ± 1.056 | 3.546 ± 1.114 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
