
Streptococcus satellite phage Javan96
Taxonomy: Viruses; unclassified bacterial viruses
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
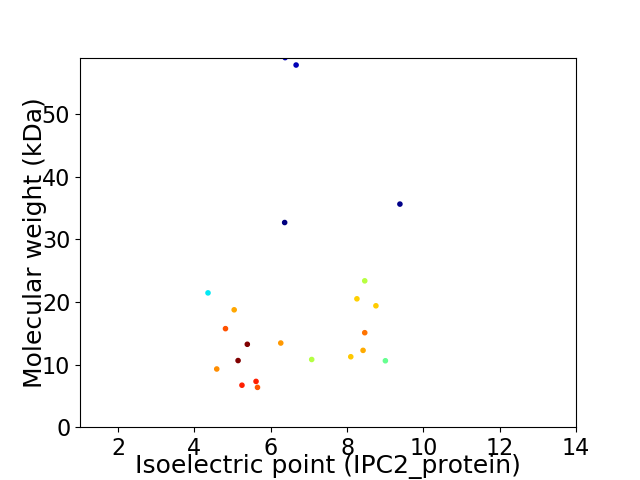
Virtual 2D-PAGE plot for 22 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
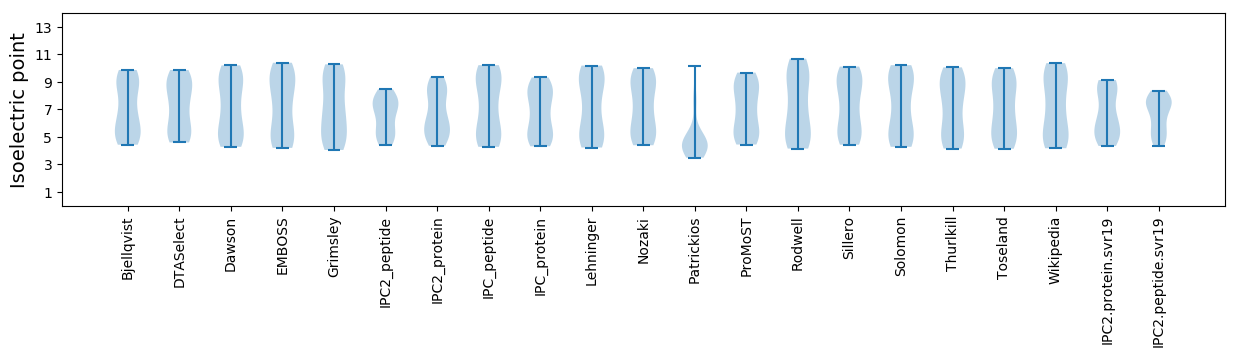
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D5ZXV3|A0A4D5ZXV3_9VIRU ATP-dependent DNA helicase OS=Streptococcus satellite phage Javan96 OX=2558868 GN=JavanS96_0002 PE=3 SV=1
MM1 pKa = 7.55RR2 pKa = 11.84TFSDD6 pKa = 3.67TPKK9 pKa = 10.21TFTFHH14 pKa = 5.7YY15 pKa = 8.55TFKK18 pKa = 11.1DD19 pKa = 3.09FDD21 pKa = 4.09TAQVACHH28 pKa = 6.88AILGYY33 pKa = 7.77MTGTYY38 pKa = 7.11EE39 pKa = 4.76QPVIDD44 pKa = 3.97ATYY47 pKa = 10.82HH48 pKa = 6.24NDD50 pKa = 3.59DD51 pKa = 3.72QGGHH55 pKa = 6.59ANQLVLEE62 pKa = 4.2YY63 pKa = 11.27AEE65 pKa = 4.49DD66 pKa = 3.69RR67 pKa = 11.84KK68 pKa = 10.66LSKK71 pKa = 10.32VFKK74 pKa = 10.15RR75 pKa = 11.84ICDD78 pKa = 3.69SFKK81 pKa = 10.93DD82 pKa = 4.21YY83 pKa = 11.52YY84 pKa = 10.51NQPEE88 pKa = 4.58DD89 pKa = 3.61MTDD92 pKa = 3.66DD93 pKa = 4.18EE94 pKa = 5.61LDD96 pKa = 3.47NMAQEE101 pKa = 4.21IEE103 pKa = 4.08LLKK106 pKa = 10.67EE107 pKa = 4.01VEE109 pKa = 4.31EE110 pKa = 5.35LDD112 pKa = 3.77DD113 pKa = 3.69QRR115 pKa = 11.84VVSLSKK121 pKa = 9.84STQEE125 pKa = 4.12KK126 pKa = 10.59VNDD129 pKa = 4.26RR130 pKa = 11.84DD131 pKa = 3.57TLMTFISDD139 pKa = 3.59HH140 pKa = 5.84NQLAEE145 pKa = 4.25HH146 pKa = 6.91LSMNYY151 pKa = 10.19KK152 pKa = 10.52EE153 pKa = 4.5MTPEE157 pKa = 3.92DD158 pKa = 3.6LGAILEE164 pKa = 4.71SISQAFNQLYY174 pKa = 10.85DD175 pKa = 3.68MVVEE179 pKa = 4.31GQLLVKK185 pKa = 10.69
MM1 pKa = 7.55RR2 pKa = 11.84TFSDD6 pKa = 3.67TPKK9 pKa = 10.21TFTFHH14 pKa = 5.7YY15 pKa = 8.55TFKK18 pKa = 11.1DD19 pKa = 3.09FDD21 pKa = 4.09TAQVACHH28 pKa = 6.88AILGYY33 pKa = 7.77MTGTYY38 pKa = 7.11EE39 pKa = 4.76QPVIDD44 pKa = 3.97ATYY47 pKa = 10.82HH48 pKa = 6.24NDD50 pKa = 3.59DD51 pKa = 3.72QGGHH55 pKa = 6.59ANQLVLEE62 pKa = 4.2YY63 pKa = 11.27AEE65 pKa = 4.49DD66 pKa = 3.69RR67 pKa = 11.84KK68 pKa = 10.66LSKK71 pKa = 10.32VFKK74 pKa = 10.15RR75 pKa = 11.84ICDD78 pKa = 3.69SFKK81 pKa = 10.93DD82 pKa = 4.21YY83 pKa = 11.52YY84 pKa = 10.51NQPEE88 pKa = 4.58DD89 pKa = 3.61MTDD92 pKa = 3.66DD93 pKa = 4.18EE94 pKa = 5.61LDD96 pKa = 3.47NMAQEE101 pKa = 4.21IEE103 pKa = 4.08LLKK106 pKa = 10.67EE107 pKa = 4.01VEE109 pKa = 4.31EE110 pKa = 5.35LDD112 pKa = 3.77DD113 pKa = 3.69QRR115 pKa = 11.84VVSLSKK121 pKa = 9.84STQEE125 pKa = 4.12KK126 pKa = 10.59VNDD129 pKa = 4.26RR130 pKa = 11.84DD131 pKa = 3.57TLMTFISDD139 pKa = 3.59HH140 pKa = 5.84NQLAEE145 pKa = 4.25HH146 pKa = 6.91LSMNYY151 pKa = 10.19KK152 pKa = 10.52EE153 pKa = 4.5MTPEE157 pKa = 3.92DD158 pKa = 3.6LGAILEE164 pKa = 4.71SISQAFNQLYY174 pKa = 10.85DD175 pKa = 3.68MVVEE179 pKa = 4.31GQLLVKK185 pKa = 10.69
Molecular weight: 21.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D6A270|A0A4D6A270_9VIRU Uncharacterized protein OS=Streptococcus satellite phage Javan96 OX=2558868 GN=JavanS96_0011 PE=4 SV=1
MM1 pKa = 7.67AEE3 pKa = 4.36LWLKK7 pKa = 10.69SYY9 pKa = 11.01QLTVKK14 pKa = 9.5PQTYY18 pKa = 10.31EE19 pKa = 3.93STKK22 pKa = 10.89LLLKK26 pKa = 10.55NHH28 pKa = 7.01ILPVFGNMKK37 pKa = 10.26LEE39 pKa = 4.9RR40 pKa = 11.84ITPSFVQQFANKK52 pKa = 9.47LAHH55 pKa = 5.94TLVNFKK61 pKa = 9.7VACSINRR68 pKa = 11.84RR69 pKa = 11.84ILQYY73 pKa = 11.24AVLLQLIPYY82 pKa = 8.96NPARR86 pKa = 11.84EE87 pKa = 3.87IIIPKK92 pKa = 8.76KK93 pKa = 9.28QKK95 pKa = 10.4KK96 pKa = 9.27SADD99 pKa = 3.3RR100 pKa = 11.84VKK102 pKa = 10.84FINPQNLKK110 pKa = 10.78ALLDD114 pKa = 3.87YY115 pKa = 10.8MEE117 pKa = 4.77TLAPTKK123 pKa = 9.68YY124 pKa = 9.8QYY126 pKa = 11.54YY127 pKa = 8.91YY128 pKa = 11.44DD129 pKa = 3.68NVLYY133 pKa = 10.56RR134 pKa = 11.84FLLATGCRR142 pKa = 11.84FGEE145 pKa = 4.29AVALEE150 pKa = 4.2WSDD153 pKa = 3.19IDD155 pKa = 5.0LEE157 pKa = 5.16AGTVSISKK165 pKa = 9.17TYY167 pKa = 10.53NRR169 pKa = 11.84QINQISTPKK178 pKa = 8.61TKK180 pKa = 10.09SGKK183 pKa = 10.17RR184 pKa = 11.84IISIDD189 pKa = 3.49NKK191 pKa = 10.74LVLLLKK197 pKa = 10.05QYY199 pKa = 11.05RR200 pKa = 11.84NRR202 pKa = 11.84QRR204 pKa = 11.84LIFIEE209 pKa = 4.41IGAHH213 pKa = 4.94APKK216 pKa = 10.57VIFASPTLTYY226 pKa = 10.59ASSDD230 pKa = 3.6VRR232 pKa = 11.84SKK234 pKa = 11.51ALAHH238 pKa = 6.06RR239 pKa = 11.84CKK241 pKa = 10.54EE242 pKa = 3.5IGIPRR247 pKa = 11.84FTFHH251 pKa = 7.74AFRR254 pKa = 11.84HH255 pKa = 4.69THH257 pKa = 6.92ASLLLNAGISYY268 pKa = 10.65KK269 pKa = 10.3EE270 pKa = 3.81LQHH273 pKa = 7.18RR274 pKa = 11.84LGHH277 pKa = 5.95SNIKK281 pKa = 7.93MTLDD285 pKa = 3.46TYY287 pKa = 11.1GHH289 pKa = 6.42ISKK292 pKa = 10.22EE293 pKa = 4.12KK294 pKa = 8.59EE295 pKa = 3.91KK296 pKa = 10.84EE297 pKa = 3.89AVSYY301 pKa = 10.0FEE303 pKa = 5.0KK304 pKa = 10.72AINNLL309 pKa = 3.44
MM1 pKa = 7.67AEE3 pKa = 4.36LWLKK7 pKa = 10.69SYY9 pKa = 11.01QLTVKK14 pKa = 9.5PQTYY18 pKa = 10.31EE19 pKa = 3.93STKK22 pKa = 10.89LLLKK26 pKa = 10.55NHH28 pKa = 7.01ILPVFGNMKK37 pKa = 10.26LEE39 pKa = 4.9RR40 pKa = 11.84ITPSFVQQFANKK52 pKa = 9.47LAHH55 pKa = 5.94TLVNFKK61 pKa = 9.7VACSINRR68 pKa = 11.84RR69 pKa = 11.84ILQYY73 pKa = 11.24AVLLQLIPYY82 pKa = 8.96NPARR86 pKa = 11.84EE87 pKa = 3.87IIIPKK92 pKa = 8.76KK93 pKa = 9.28QKK95 pKa = 10.4KK96 pKa = 9.27SADD99 pKa = 3.3RR100 pKa = 11.84VKK102 pKa = 10.84FINPQNLKK110 pKa = 10.78ALLDD114 pKa = 3.87YY115 pKa = 10.8MEE117 pKa = 4.77TLAPTKK123 pKa = 9.68YY124 pKa = 9.8QYY126 pKa = 11.54YY127 pKa = 8.91YY128 pKa = 11.44DD129 pKa = 3.68NVLYY133 pKa = 10.56RR134 pKa = 11.84FLLATGCRR142 pKa = 11.84FGEE145 pKa = 4.29AVALEE150 pKa = 4.2WSDD153 pKa = 3.19IDD155 pKa = 5.0LEE157 pKa = 5.16AGTVSISKK165 pKa = 9.17TYY167 pKa = 10.53NRR169 pKa = 11.84QINQISTPKK178 pKa = 8.61TKK180 pKa = 10.09SGKK183 pKa = 10.17RR184 pKa = 11.84IISIDD189 pKa = 3.49NKK191 pKa = 10.74LVLLLKK197 pKa = 10.05QYY199 pKa = 11.05RR200 pKa = 11.84NRR202 pKa = 11.84QRR204 pKa = 11.84LIFIEE209 pKa = 4.41IGAHH213 pKa = 4.94APKK216 pKa = 10.57VIFASPTLTYY226 pKa = 10.59ASSDD230 pKa = 3.6VRR232 pKa = 11.84SKK234 pKa = 11.51ALAHH238 pKa = 6.06RR239 pKa = 11.84CKK241 pKa = 10.54EE242 pKa = 3.5IGIPRR247 pKa = 11.84FTFHH251 pKa = 7.74AFRR254 pKa = 11.84HH255 pKa = 4.69THH257 pKa = 6.92ASLLLNAGISYY268 pKa = 10.65KK269 pKa = 10.3EE270 pKa = 3.81LQHH273 pKa = 7.18RR274 pKa = 11.84LGHH277 pKa = 5.95SNIKK281 pKa = 7.93MTLDD285 pKa = 3.46TYY287 pKa = 11.1GHH289 pKa = 6.42ISKK292 pKa = 10.22EE293 pKa = 4.12KK294 pKa = 8.59EE295 pKa = 3.91KK296 pKa = 10.84EE297 pKa = 3.89AVSYY301 pKa = 10.0FEE303 pKa = 5.0KK304 pKa = 10.72AINNLL309 pKa = 3.44
Molecular weight: 35.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3716 |
57 |
504 |
168.9 |
19.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.705 ± 0.415 | 0.78 ± 0.189 |
5.813 ± 0.465 | 8.073 ± 0.397 |
4.36 ± 0.248 | 4.413 ± 0.284 |
2.045 ± 0.251 | 7.481 ± 0.437 |
8.665 ± 0.4 | 10.495 ± 0.493 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.153 ± 0.266 | 5.84 ± 0.532 |
3.498 ± 0.472 | 4.171 ± 0.403 |
5.328 ± 0.276 | 5.274 ± 0.399 |
5.705 ± 0.285 | 4.736 ± 0.359 |
0.942 ± 0.153 | 4.521 ± 0.342 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
