
Mycoplasma genitalium (strain ATCC 33530 / G-37 / NCTC 10195)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Tenericutes; Mollicutes; Mycoplasmatales; Mycoplasmataceae; Mycoplasma; Mycoplasma genitalium
Average proteome isoelectric point is 7.94
Get precalculated fractions of proteins
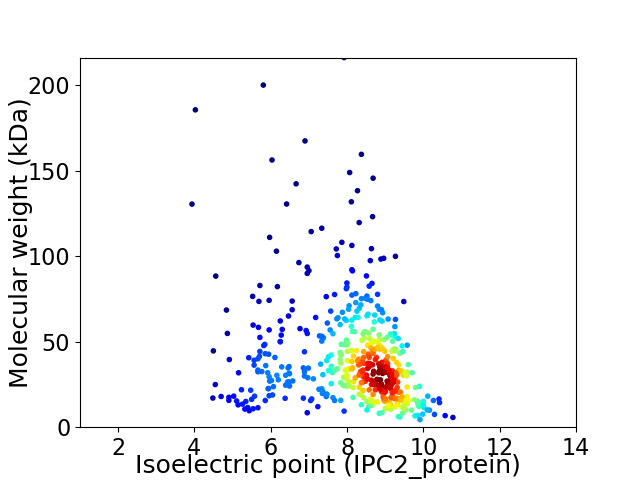
Virtual 2D-PAGE plot for 483 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q49416|Y315_MYCGE Uncharacterized protein MG315 OS=Mycoplasma genitalium (strain ATCC 33530 / G-37 / NCTC 10195) OX=243273 GN=MG315 PE=4 SV=1
MM1 pKa = 7.8AKK3 pKa = 10.05NKK5 pKa = 10.1QSVFEE10 pKa = 4.33EE11 pKa = 4.06KK12 pKa = 10.62NYY14 pKa = 9.16TQTEE18 pKa = 4.21PEE20 pKa = 4.46NIFGDD25 pKa = 4.13LYY27 pKa = 10.96DD28 pKa = 4.14GKK30 pKa = 9.92STVEE34 pKa = 3.81EE35 pKa = 4.29DD36 pKa = 3.62PNIKK40 pKa = 9.73VAYY43 pKa = 9.7DD44 pKa = 3.48ADD46 pKa = 3.92GNGYY50 pKa = 10.23YY51 pKa = 9.77IAFNKK56 pKa = 7.51EE57 pKa = 3.54TGVYY61 pKa = 8.99YY62 pKa = 10.46DD63 pKa = 4.21PYY65 pKa = 11.39GDD67 pKa = 3.73TEE69 pKa = 4.27YY70 pKa = 10.98DD71 pKa = 2.89ISQLFDD77 pKa = 3.55EE78 pKa = 5.14NGNPFVFDD86 pKa = 3.98EE87 pKa = 4.32KK88 pKa = 11.11QEE90 pKa = 4.01EE91 pKa = 4.06NDD93 pKa = 3.24YY94 pKa = 11.22LKK96 pKa = 11.13YY97 pKa = 10.76VGNPDD102 pKa = 3.29YY103 pKa = 11.33GSYY106 pKa = 10.8DD107 pKa = 3.73EE108 pKa = 4.59NGEE111 pKa = 4.44WVWSGYY117 pKa = 9.4FEE119 pKa = 4.58NDD121 pKa = 2.39QWISTKK127 pKa = 10.17EE128 pKa = 3.94SQPTDD133 pKa = 3.12EE134 pKa = 5.56NYY136 pKa = 10.95GFDD139 pKa = 3.5SDD141 pKa = 5.25LPPEE145 pKa = 4.45VKK147 pKa = 10.3QPEE150 pKa = 4.54SVEE153 pKa = 4.07DD154 pKa = 3.97NYY156 pKa = 11.61GFDD159 pKa = 3.62NDD161 pKa = 4.66LPPEE165 pKa = 4.32VKK167 pKa = 10.29QPEE170 pKa = 4.54SVEE173 pKa = 4.07DD174 pKa = 3.97NYY176 pKa = 11.61GFDD179 pKa = 3.62NDD181 pKa = 4.66LPPEE185 pKa = 4.32VKK187 pKa = 10.33QPEE190 pKa = 4.56SVVDD194 pKa = 3.83QPSSDD199 pKa = 4.5DD200 pKa = 3.74YY201 pKa = 11.12FAKK204 pKa = 10.55QPTDD208 pKa = 3.07EE209 pKa = 5.49NYY211 pKa = 10.95GFDD214 pKa = 3.6NDD216 pKa = 4.58LPPEE220 pKa = 4.32VKK222 pKa = 10.33QPEE225 pKa = 4.56SVVDD229 pKa = 3.81QPSSDD234 pKa = 3.55DD235 pKa = 3.91HH236 pKa = 6.48FAKK239 pKa = 10.46QPEE242 pKa = 4.67STTDD246 pKa = 3.06SYY248 pKa = 12.08SFDD251 pKa = 3.63SDD253 pKa = 4.32LPQPTLDD260 pKa = 3.76QPSLDD265 pKa = 4.04DD266 pKa = 3.59HH267 pKa = 6.0VQYY270 pKa = 11.32NFDD273 pKa = 3.2HH274 pKa = 7.15HH275 pKa = 7.07EE276 pKa = 3.93EE277 pKa = 4.08LKK279 pKa = 10.82PVAEE283 pKa = 4.32EE284 pKa = 3.74QNNYY288 pKa = 7.85QVGFDD293 pKa = 3.48QVQANLDD300 pKa = 3.89NNEE303 pKa = 4.37EE304 pKa = 4.04IQPTAEE310 pKa = 4.33KK311 pKa = 10.63KK312 pKa = 8.8VTTDD316 pKa = 4.07FEE318 pKa = 4.83SKK320 pKa = 9.39QAQVVDD326 pKa = 4.67SYY328 pKa = 11.47QLPIDD333 pKa = 4.43TDD335 pKa = 3.66QQDD338 pKa = 3.22QTTFSSSFEE347 pKa = 4.09TQPTVEE353 pKa = 5.2QFDD356 pKa = 3.93QVNSEE361 pKa = 4.27VNDD364 pKa = 3.4QFKK367 pKa = 11.16PEE369 pKa = 3.92ITKK372 pKa = 10.68EE373 pKa = 3.99PVLEE377 pKa = 4.19SSFNKK382 pKa = 9.67QDD384 pKa = 3.46VVEE387 pKa = 4.26TSDD390 pKa = 5.04LNSEE394 pKa = 4.35SNLYY398 pKa = 10.44SEE400 pKa = 4.7NNKK403 pKa = 10.25DD404 pKa = 3.22ATNNDD409 pKa = 3.48SLNSEE414 pKa = 5.14FIQLNSNSEE423 pKa = 4.05TASDD427 pKa = 3.72DD428 pKa = 3.2VHH430 pKa = 7.83YY431 pKa = 9.98EE432 pKa = 4.18SKK434 pKa = 10.7SEE436 pKa = 4.51PIHH439 pKa = 6.89DD440 pKa = 3.96YY441 pKa = 11.35KK442 pKa = 11.08FGSDD446 pKa = 3.38LSQSNSNNSLEE457 pKa = 4.13SEE459 pKa = 4.01PVKK462 pKa = 10.75FNSEE466 pKa = 4.23TAPDD470 pKa = 3.34AHH472 pKa = 6.94FEE474 pKa = 4.4SQSEE478 pKa = 4.18PVDD481 pKa = 3.38QVQYY485 pKa = 10.96DD486 pKa = 3.8IYY488 pKa = 11.12QNEE491 pKa = 4.23EE492 pKa = 4.14LKK494 pKa = 9.5PTLDD498 pKa = 3.66QPSSDD503 pKa = 4.97DD504 pKa = 3.73YY505 pKa = 11.12FAKK508 pKa = 10.55QPTDD512 pKa = 3.07EE513 pKa = 5.49NYY515 pKa = 10.95GFDD518 pKa = 3.6NDD520 pKa = 4.58LPPEE524 pKa = 4.32VKK526 pKa = 10.33QPEE529 pKa = 4.56SVVDD533 pKa = 3.81QPSSDD538 pKa = 3.55DD539 pKa = 3.91HH540 pKa = 6.48FAKK543 pKa = 10.46QPEE546 pKa = 4.67STTDD550 pKa = 3.06SYY552 pKa = 12.08SFDD555 pKa = 3.63SDD557 pKa = 4.32LPQPTLDD564 pKa = 3.76QPSLDD569 pKa = 4.04DD570 pKa = 3.59HH571 pKa = 6.0VQYY574 pKa = 11.32NFDD577 pKa = 3.2HH578 pKa = 7.15HH579 pKa = 7.07EE580 pKa = 3.93EE581 pKa = 4.08LKK583 pKa = 10.82PVAEE587 pKa = 4.32EE588 pKa = 3.74QNNYY592 pKa = 7.85QVGFDD597 pKa = 3.48QVQANLDD604 pKa = 3.89NNEE607 pKa = 4.37EE608 pKa = 4.04IQPTAEE614 pKa = 4.42KK615 pKa = 10.55EE616 pKa = 4.16VTTDD620 pKa = 4.16FEE622 pKa = 4.82SKK624 pKa = 9.39QAQVVDD630 pKa = 4.67SYY632 pKa = 11.47QLPIDD637 pKa = 4.43TDD639 pKa = 3.66QQDD642 pKa = 3.22QTTFSSSFEE651 pKa = 4.09TQPTVEE657 pKa = 5.2QFDD660 pKa = 3.93QVNSEE665 pKa = 4.27VNDD668 pKa = 3.4QFKK671 pKa = 11.16PEE673 pKa = 3.92ITKK676 pKa = 10.68EE677 pKa = 3.99PVLEE681 pKa = 4.19SSFNKK686 pKa = 9.67QDD688 pKa = 3.4VVEE691 pKa = 4.29TSNYY695 pKa = 8.6TNNLQKK701 pKa = 10.58FDD703 pKa = 3.65IQSDD707 pKa = 3.67NKK709 pKa = 9.66ITITTKK715 pKa = 10.32KK716 pKa = 9.98SSPQIPTTLPISFVSNRR733 pKa = 11.84IEE735 pKa = 4.09YY736 pKa = 10.5KK737 pKa = 10.26PVEE740 pKa = 4.31TLALDD745 pKa = 3.74NKK747 pKa = 10.22EE748 pKa = 4.17SQQEE752 pKa = 4.04QITINSITEE761 pKa = 3.93DD762 pKa = 3.6SKK764 pKa = 10.29TLAKK768 pKa = 9.54TLSVQLQQINSLNNQSIVTSEE789 pKa = 4.16SVRR792 pKa = 11.84LDD794 pKa = 3.46KK795 pKa = 11.16KK796 pKa = 10.91DD797 pKa = 3.64DD798 pKa = 3.5QLTINTVNSEE808 pKa = 4.0DD809 pKa = 3.73QQPKK813 pKa = 9.92IEE815 pKa = 4.33VFVKK819 pKa = 10.42AKK821 pKa = 10.5EE822 pKa = 3.97PVEE825 pKa = 4.03EE826 pKa = 5.14HH827 pKa = 7.21SITQNKK833 pKa = 8.69QSVEE837 pKa = 4.21DD838 pKa = 4.46KK839 pKa = 11.42SEE841 pKa = 4.0LDD843 pKa = 3.55NFNKK847 pKa = 10.34KK848 pKa = 10.01SDD850 pKa = 3.47LYY852 pKa = 11.22KK853 pKa = 10.38IISEE857 pKa = 4.37LKK859 pKa = 10.06RR860 pKa = 11.84GEE862 pKa = 4.32LNPTINFDD870 pKa = 6.18AIFQMNDD877 pKa = 2.64YY878 pKa = 10.68QMSVKK883 pKa = 10.34QSFIHH888 pKa = 7.09LNDD891 pKa = 3.29FVTNYY896 pKa = 10.1KK897 pKa = 9.89NQISEE902 pKa = 4.25RR903 pKa = 11.84YY904 pKa = 9.6LIIKK908 pKa = 10.18KK909 pKa = 9.39EE910 pKa = 4.04LQSEE914 pKa = 4.63LSRR917 pKa = 11.84LIDD920 pKa = 3.37QNEE923 pKa = 4.01NLNVQFNNAKK933 pKa = 10.3NLTTLQKK940 pKa = 11.17EE941 pKa = 4.13EE942 pKa = 4.52MIRR945 pKa = 11.84SLASDD950 pKa = 3.51FAIAYY955 pKa = 7.96KK956 pKa = 10.34PSNSYY961 pKa = 10.23EE962 pKa = 3.94QLQKK966 pKa = 10.5SGEE969 pKa = 4.2IMRR972 pKa = 11.84HH973 pKa = 3.74VQRR976 pKa = 11.84AITEE980 pKa = 4.08NEE982 pKa = 3.92KK983 pKa = 10.72KK984 pKa = 10.08IEE986 pKa = 4.45SIQGSLKK993 pKa = 9.69QLKK996 pKa = 7.76TVYY999 pKa = 10.18NSCCEE1004 pKa = 4.09TIMNNINKK1012 pKa = 9.56LDD1014 pKa = 3.6NTLRR1018 pKa = 11.84FAKK1021 pKa = 10.23KK1022 pKa = 10.24EE1023 pKa = 3.85KK1024 pKa = 10.76DD1025 pKa = 3.33PLLLSNFDD1033 pKa = 3.85SVTDD1037 pKa = 3.71NGLVEE1042 pKa = 4.81PNQLMDD1048 pKa = 5.06DD1049 pKa = 5.21LIDD1052 pKa = 4.19FSNTFDD1058 pKa = 4.49NISNEE1063 pKa = 3.85QLDD1066 pKa = 3.87DD1067 pKa = 4.77FIYY1070 pKa = 11.06EE1071 pKa = 4.07NMDD1074 pKa = 3.5RR1075 pKa = 11.84NIDD1078 pKa = 3.74FEE1080 pKa = 5.18FEE1082 pKa = 3.97GFNNDD1087 pKa = 4.72FVDD1090 pKa = 3.62IDD1092 pKa = 4.19AKK1094 pKa = 11.61VMDD1097 pKa = 4.32SMSAFSVNDD1106 pKa = 3.61LDD1108 pKa = 5.67IEE1110 pKa = 4.44TLVPDD1115 pKa = 3.65RR1116 pKa = 11.84TSNFSSLLDD1125 pKa = 3.59EE1126 pKa = 5.74DD1127 pKa = 5.16LFEE1130 pKa = 5.41SSGDD1134 pKa = 3.6FSLDD1138 pKa = 3.42YY1139 pKa = 11.31
MM1 pKa = 7.8AKK3 pKa = 10.05NKK5 pKa = 10.1QSVFEE10 pKa = 4.33EE11 pKa = 4.06KK12 pKa = 10.62NYY14 pKa = 9.16TQTEE18 pKa = 4.21PEE20 pKa = 4.46NIFGDD25 pKa = 4.13LYY27 pKa = 10.96DD28 pKa = 4.14GKK30 pKa = 9.92STVEE34 pKa = 3.81EE35 pKa = 4.29DD36 pKa = 3.62PNIKK40 pKa = 9.73VAYY43 pKa = 9.7DD44 pKa = 3.48ADD46 pKa = 3.92GNGYY50 pKa = 10.23YY51 pKa = 9.77IAFNKK56 pKa = 7.51EE57 pKa = 3.54TGVYY61 pKa = 8.99YY62 pKa = 10.46DD63 pKa = 4.21PYY65 pKa = 11.39GDD67 pKa = 3.73TEE69 pKa = 4.27YY70 pKa = 10.98DD71 pKa = 2.89ISQLFDD77 pKa = 3.55EE78 pKa = 5.14NGNPFVFDD86 pKa = 3.98EE87 pKa = 4.32KK88 pKa = 11.11QEE90 pKa = 4.01EE91 pKa = 4.06NDD93 pKa = 3.24YY94 pKa = 11.22LKK96 pKa = 11.13YY97 pKa = 10.76VGNPDD102 pKa = 3.29YY103 pKa = 11.33GSYY106 pKa = 10.8DD107 pKa = 3.73EE108 pKa = 4.59NGEE111 pKa = 4.44WVWSGYY117 pKa = 9.4FEE119 pKa = 4.58NDD121 pKa = 2.39QWISTKK127 pKa = 10.17EE128 pKa = 3.94SQPTDD133 pKa = 3.12EE134 pKa = 5.56NYY136 pKa = 10.95GFDD139 pKa = 3.5SDD141 pKa = 5.25LPPEE145 pKa = 4.45VKK147 pKa = 10.3QPEE150 pKa = 4.54SVEE153 pKa = 4.07DD154 pKa = 3.97NYY156 pKa = 11.61GFDD159 pKa = 3.62NDD161 pKa = 4.66LPPEE165 pKa = 4.32VKK167 pKa = 10.29QPEE170 pKa = 4.54SVEE173 pKa = 4.07DD174 pKa = 3.97NYY176 pKa = 11.61GFDD179 pKa = 3.62NDD181 pKa = 4.66LPPEE185 pKa = 4.32VKK187 pKa = 10.33QPEE190 pKa = 4.56SVVDD194 pKa = 3.83QPSSDD199 pKa = 4.5DD200 pKa = 3.74YY201 pKa = 11.12FAKK204 pKa = 10.55QPTDD208 pKa = 3.07EE209 pKa = 5.49NYY211 pKa = 10.95GFDD214 pKa = 3.6NDD216 pKa = 4.58LPPEE220 pKa = 4.32VKK222 pKa = 10.33QPEE225 pKa = 4.56SVVDD229 pKa = 3.81QPSSDD234 pKa = 3.55DD235 pKa = 3.91HH236 pKa = 6.48FAKK239 pKa = 10.46QPEE242 pKa = 4.67STTDD246 pKa = 3.06SYY248 pKa = 12.08SFDD251 pKa = 3.63SDD253 pKa = 4.32LPQPTLDD260 pKa = 3.76QPSLDD265 pKa = 4.04DD266 pKa = 3.59HH267 pKa = 6.0VQYY270 pKa = 11.32NFDD273 pKa = 3.2HH274 pKa = 7.15HH275 pKa = 7.07EE276 pKa = 3.93EE277 pKa = 4.08LKK279 pKa = 10.82PVAEE283 pKa = 4.32EE284 pKa = 3.74QNNYY288 pKa = 7.85QVGFDD293 pKa = 3.48QVQANLDD300 pKa = 3.89NNEE303 pKa = 4.37EE304 pKa = 4.04IQPTAEE310 pKa = 4.33KK311 pKa = 10.63KK312 pKa = 8.8VTTDD316 pKa = 4.07FEE318 pKa = 4.83SKK320 pKa = 9.39QAQVVDD326 pKa = 4.67SYY328 pKa = 11.47QLPIDD333 pKa = 4.43TDD335 pKa = 3.66QQDD338 pKa = 3.22QTTFSSSFEE347 pKa = 4.09TQPTVEE353 pKa = 5.2QFDD356 pKa = 3.93QVNSEE361 pKa = 4.27VNDD364 pKa = 3.4QFKK367 pKa = 11.16PEE369 pKa = 3.92ITKK372 pKa = 10.68EE373 pKa = 3.99PVLEE377 pKa = 4.19SSFNKK382 pKa = 9.67QDD384 pKa = 3.46VVEE387 pKa = 4.26TSDD390 pKa = 5.04LNSEE394 pKa = 4.35SNLYY398 pKa = 10.44SEE400 pKa = 4.7NNKK403 pKa = 10.25DD404 pKa = 3.22ATNNDD409 pKa = 3.48SLNSEE414 pKa = 5.14FIQLNSNSEE423 pKa = 4.05TASDD427 pKa = 3.72DD428 pKa = 3.2VHH430 pKa = 7.83YY431 pKa = 9.98EE432 pKa = 4.18SKK434 pKa = 10.7SEE436 pKa = 4.51PIHH439 pKa = 6.89DD440 pKa = 3.96YY441 pKa = 11.35KK442 pKa = 11.08FGSDD446 pKa = 3.38LSQSNSNNSLEE457 pKa = 4.13SEE459 pKa = 4.01PVKK462 pKa = 10.75FNSEE466 pKa = 4.23TAPDD470 pKa = 3.34AHH472 pKa = 6.94FEE474 pKa = 4.4SQSEE478 pKa = 4.18PVDD481 pKa = 3.38QVQYY485 pKa = 10.96DD486 pKa = 3.8IYY488 pKa = 11.12QNEE491 pKa = 4.23EE492 pKa = 4.14LKK494 pKa = 9.5PTLDD498 pKa = 3.66QPSSDD503 pKa = 4.97DD504 pKa = 3.73YY505 pKa = 11.12FAKK508 pKa = 10.55QPTDD512 pKa = 3.07EE513 pKa = 5.49NYY515 pKa = 10.95GFDD518 pKa = 3.6NDD520 pKa = 4.58LPPEE524 pKa = 4.32VKK526 pKa = 10.33QPEE529 pKa = 4.56SVVDD533 pKa = 3.81QPSSDD538 pKa = 3.55DD539 pKa = 3.91HH540 pKa = 6.48FAKK543 pKa = 10.46QPEE546 pKa = 4.67STTDD550 pKa = 3.06SYY552 pKa = 12.08SFDD555 pKa = 3.63SDD557 pKa = 4.32LPQPTLDD564 pKa = 3.76QPSLDD569 pKa = 4.04DD570 pKa = 3.59HH571 pKa = 6.0VQYY574 pKa = 11.32NFDD577 pKa = 3.2HH578 pKa = 7.15HH579 pKa = 7.07EE580 pKa = 3.93EE581 pKa = 4.08LKK583 pKa = 10.82PVAEE587 pKa = 4.32EE588 pKa = 3.74QNNYY592 pKa = 7.85QVGFDD597 pKa = 3.48QVQANLDD604 pKa = 3.89NNEE607 pKa = 4.37EE608 pKa = 4.04IQPTAEE614 pKa = 4.42KK615 pKa = 10.55EE616 pKa = 4.16VTTDD620 pKa = 4.16FEE622 pKa = 4.82SKK624 pKa = 9.39QAQVVDD630 pKa = 4.67SYY632 pKa = 11.47QLPIDD637 pKa = 4.43TDD639 pKa = 3.66QQDD642 pKa = 3.22QTTFSSSFEE651 pKa = 4.09TQPTVEE657 pKa = 5.2QFDD660 pKa = 3.93QVNSEE665 pKa = 4.27VNDD668 pKa = 3.4QFKK671 pKa = 11.16PEE673 pKa = 3.92ITKK676 pKa = 10.68EE677 pKa = 3.99PVLEE681 pKa = 4.19SSFNKK686 pKa = 9.67QDD688 pKa = 3.4VVEE691 pKa = 4.29TSNYY695 pKa = 8.6TNNLQKK701 pKa = 10.58FDD703 pKa = 3.65IQSDD707 pKa = 3.67NKK709 pKa = 9.66ITITTKK715 pKa = 10.32KK716 pKa = 9.98SSPQIPTTLPISFVSNRR733 pKa = 11.84IEE735 pKa = 4.09YY736 pKa = 10.5KK737 pKa = 10.26PVEE740 pKa = 4.31TLALDD745 pKa = 3.74NKK747 pKa = 10.22EE748 pKa = 4.17SQQEE752 pKa = 4.04QITINSITEE761 pKa = 3.93DD762 pKa = 3.6SKK764 pKa = 10.29TLAKK768 pKa = 9.54TLSVQLQQINSLNNQSIVTSEE789 pKa = 4.16SVRR792 pKa = 11.84LDD794 pKa = 3.46KK795 pKa = 11.16KK796 pKa = 10.91DD797 pKa = 3.64DD798 pKa = 3.5QLTINTVNSEE808 pKa = 4.0DD809 pKa = 3.73QQPKK813 pKa = 9.92IEE815 pKa = 4.33VFVKK819 pKa = 10.42AKK821 pKa = 10.5EE822 pKa = 3.97PVEE825 pKa = 4.03EE826 pKa = 5.14HH827 pKa = 7.21SITQNKK833 pKa = 8.69QSVEE837 pKa = 4.21DD838 pKa = 4.46KK839 pKa = 11.42SEE841 pKa = 4.0LDD843 pKa = 3.55NFNKK847 pKa = 10.34KK848 pKa = 10.01SDD850 pKa = 3.47LYY852 pKa = 11.22KK853 pKa = 10.38IISEE857 pKa = 4.37LKK859 pKa = 10.06RR860 pKa = 11.84GEE862 pKa = 4.32LNPTINFDD870 pKa = 6.18AIFQMNDD877 pKa = 2.64YY878 pKa = 10.68QMSVKK883 pKa = 10.34QSFIHH888 pKa = 7.09LNDD891 pKa = 3.29FVTNYY896 pKa = 10.1KK897 pKa = 9.89NQISEE902 pKa = 4.25RR903 pKa = 11.84YY904 pKa = 9.6LIIKK908 pKa = 10.18KK909 pKa = 9.39EE910 pKa = 4.04LQSEE914 pKa = 4.63LSRR917 pKa = 11.84LIDD920 pKa = 3.37QNEE923 pKa = 4.01NLNVQFNNAKK933 pKa = 10.3NLTTLQKK940 pKa = 11.17EE941 pKa = 4.13EE942 pKa = 4.52MIRR945 pKa = 11.84SLASDD950 pKa = 3.51FAIAYY955 pKa = 7.96KK956 pKa = 10.34PSNSYY961 pKa = 10.23EE962 pKa = 3.94QLQKK966 pKa = 10.5SGEE969 pKa = 4.2IMRR972 pKa = 11.84HH973 pKa = 3.74VQRR976 pKa = 11.84AITEE980 pKa = 4.08NEE982 pKa = 3.92KK983 pKa = 10.72KK984 pKa = 10.08IEE986 pKa = 4.45SIQGSLKK993 pKa = 9.69QLKK996 pKa = 7.76TVYY999 pKa = 10.18NSCCEE1004 pKa = 4.09TIMNNINKK1012 pKa = 9.56LDD1014 pKa = 3.6NTLRR1018 pKa = 11.84FAKK1021 pKa = 10.23KK1022 pKa = 10.24EE1023 pKa = 3.85KK1024 pKa = 10.76DD1025 pKa = 3.33PLLLSNFDD1033 pKa = 3.85SVTDD1037 pKa = 3.71NGLVEE1042 pKa = 4.81PNQLMDD1048 pKa = 5.06DD1049 pKa = 5.21LIDD1052 pKa = 4.19FSNTFDD1058 pKa = 4.49NISNEE1063 pKa = 3.85QLDD1066 pKa = 3.87DD1067 pKa = 4.77FIYY1070 pKa = 11.06EE1071 pKa = 4.07NMDD1074 pKa = 3.5RR1075 pKa = 11.84NIDD1078 pKa = 3.74FEE1080 pKa = 5.18FEE1082 pKa = 3.97GFNNDD1087 pKa = 4.72FVDD1090 pKa = 3.62IDD1092 pKa = 4.19AKK1094 pKa = 11.61VMDD1097 pKa = 4.32SMSAFSVNDD1106 pKa = 3.61LDD1108 pKa = 5.67IEE1110 pKa = 4.44TLVPDD1115 pKa = 3.65RR1116 pKa = 11.84TSNFSSLLDD1125 pKa = 3.59EE1126 pKa = 5.74DD1127 pKa = 5.16LFEE1130 pKa = 5.41SSGDD1134 pKa = 3.6FSLDD1138 pKa = 3.42YY1139 pKa = 11.31
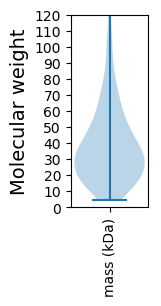
Molecular weight: 130.53 kDa
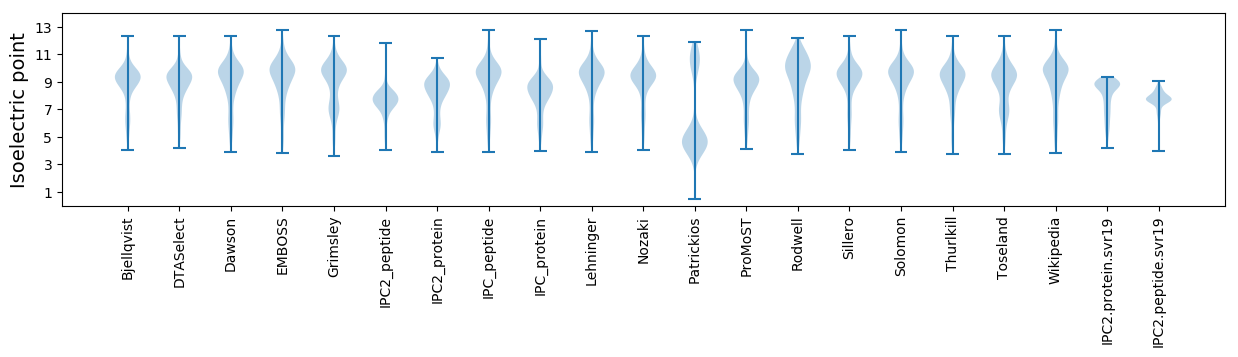
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P47414|RS5_MYCGE 30S ribosomal protein S5 OS=Mycoplasma genitalium (strain ATCC 33530 / G-37 / NCTC 10195) OX=243273 GN=rpsE PE=3 SV=1
MM1 pKa = 7.61AKK3 pKa = 10.14KK4 pKa = 10.1SLKK7 pKa = 10.15VKK9 pKa = 9.97QSRR12 pKa = 11.84PNKK15 pKa = 9.88FSVRR19 pKa = 11.84DD20 pKa = 3.43YY21 pKa = 10.38TRR23 pKa = 11.84CLRR26 pKa = 11.84CGRR29 pKa = 11.84ARR31 pKa = 11.84AVLSHH36 pKa = 6.81FGVCRR41 pKa = 11.84LCFRR45 pKa = 11.84EE46 pKa = 3.97LAYY49 pKa = 10.57AGAIPGVKK57 pKa = 9.35KK58 pKa = 10.94ASWW61 pKa = 3.03
MM1 pKa = 7.61AKK3 pKa = 10.14KK4 pKa = 10.1SLKK7 pKa = 10.15VKK9 pKa = 9.97QSRR12 pKa = 11.84PNKK15 pKa = 9.88FSVRR19 pKa = 11.84DD20 pKa = 3.43YY21 pKa = 10.38TRR23 pKa = 11.84CLRR26 pKa = 11.84CGRR29 pKa = 11.84ARR31 pKa = 11.84AVLSHH36 pKa = 6.81FGVCRR41 pKa = 11.84LCFRR45 pKa = 11.84EE46 pKa = 3.97LAYY49 pKa = 10.57AGAIPGVKK57 pKa = 9.35KK58 pKa = 10.94ASWW61 pKa = 3.03
Molecular weight: 6.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
177065 |
37 |
1805 |
366.6 |
41.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.566 ± 0.083 | 0.825 ± 0.036 |
4.915 ± 0.081 | 5.64 ± 0.138 |
6.158 ± 0.107 | 4.626 ± 0.095 |
1.576 ± 0.049 | 8.261 ± 0.125 |
9.502 ± 0.139 | 10.684 ± 0.137 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.528 ± 0.041 | 7.523 ± 0.116 |
2.989 ± 0.087 | 4.736 ± 0.106 |
3.092 ± 0.075 | 6.658 ± 0.116 |
5.397 ± 0.079 | 6.11 ± 0.098 |
0.972 ± 0.038 | 3.241 ± 0.05 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
