
Borrelia burgdorferi (strain ATCC 35210 / B31 / CIP 102532 / DSM 4680)
Taxonomy: cellular organisms; Bacteria; Spirochaetes; Spirochaetia; Spirochaetales; Borreliaceae; Borreliella; Borreliella burgdorferi
Average proteome isoelectric point is 7.44
Get precalculated fractions of proteins
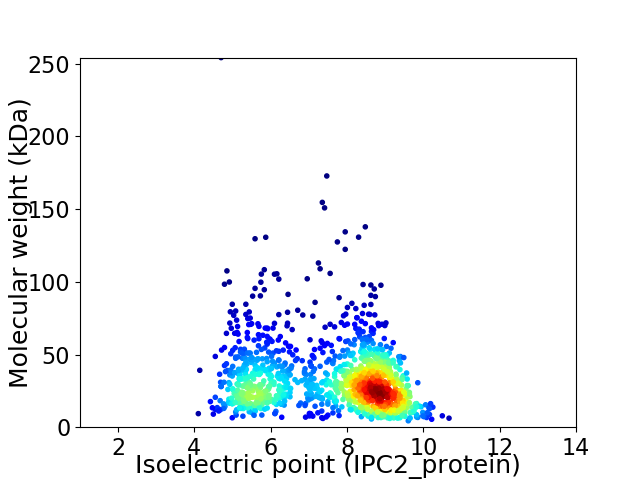
Virtual 2D-PAGE plot for 1290 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|O51298|O51298_BORBU Exported protein OS=Borrelia burgdorferi (strain ATCC 35210 / B31 / CIP 102532 / DSM 4680) OX=224326 GN=BB_0319 PE=3 SV=1
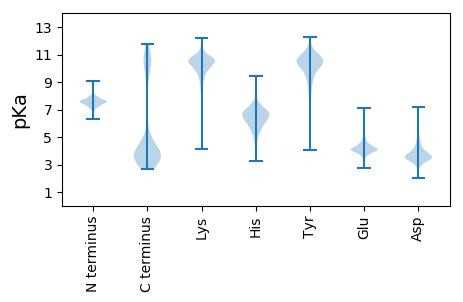
MM1 pKa = 7.57KK2 pKa = 10.5GFLAIKK8 pKa = 10.18LSNNEE13 pKa = 4.05YY14 pKa = 10.85YY15 pKa = 9.92PVLNLDD21 pKa = 3.76DD22 pKa = 5.34SSVKK26 pKa = 10.35RR27 pKa = 11.84IILGKK32 pKa = 10.04VAGDD36 pKa = 3.27TSIKK40 pKa = 10.37LNLYY44 pKa = 8.5VSEE47 pKa = 4.84YY48 pKa = 10.96EE49 pKa = 4.78DD50 pKa = 3.89FLDD53 pKa = 4.11PFLVGSFFLDD63 pKa = 3.45NLKK66 pKa = 10.63KK67 pKa = 10.64EE68 pKa = 4.3SLSVNVYY75 pKa = 9.42FKK77 pKa = 11.24VVDD80 pKa = 4.6EE81 pKa = 4.05ILYY84 pKa = 10.73VYY86 pKa = 10.62GEE88 pKa = 4.12SDD90 pKa = 4.62GIQDD94 pKa = 3.58KK95 pKa = 11.51SKK97 pKa = 11.09FDD99 pKa = 3.88LSLVDD104 pKa = 5.2FNQNLKK110 pKa = 10.55GSNRR114 pKa = 11.84GNPLNDD120 pKa = 3.7FKK122 pKa = 11.75SDD124 pKa = 3.17IGSDD128 pKa = 3.49LSVEE132 pKa = 4.18NLGSEE137 pKa = 4.07FDD139 pKa = 4.35LNFEE143 pKa = 4.34EE144 pKa = 6.32KK145 pKa = 10.74DD146 pKa = 3.65DD147 pKa = 4.0FVKK150 pKa = 10.78KK151 pKa = 10.45DD152 pKa = 3.51VSDD155 pKa = 3.77VSMPTINSLDD165 pKa = 3.71LKK167 pKa = 10.13TEE169 pKa = 3.93EE170 pKa = 4.24DD171 pKa = 3.61QKK173 pKa = 11.27IEE175 pKa = 4.3TNVSDD180 pKa = 6.53FEE182 pKa = 5.55DD183 pKa = 4.65FDD185 pKa = 6.05DD186 pKa = 5.67NYY188 pKa = 11.51DD189 pKa = 3.95DD190 pKa = 5.5SLVDD194 pKa = 3.79SKK196 pKa = 11.61KK197 pKa = 11.11LNLEE201 pKa = 4.1NKK203 pKa = 8.42KK204 pKa = 10.32VEE206 pKa = 4.38YY207 pKa = 9.94IDD209 pKa = 4.45EE210 pKa = 4.54KK211 pKa = 11.38SLNLNNFEE219 pKa = 4.04FQKK222 pKa = 10.97LEE224 pKa = 3.94EE225 pKa = 4.42SSLEE229 pKa = 4.05EE230 pKa = 4.19EE231 pKa = 4.68DD232 pKa = 4.73LHH234 pKa = 7.74DD235 pKa = 5.91LSVEE239 pKa = 4.62DD240 pKa = 5.46IIADD244 pKa = 3.4IDD246 pKa = 3.78KK247 pKa = 10.83EE248 pKa = 4.44LIDD251 pKa = 4.04TSDD254 pKa = 3.54EE255 pKa = 4.18DD256 pKa = 3.87VEE258 pKa = 5.07LDD260 pKa = 4.11LRR262 pKa = 11.84EE263 pKa = 4.59DD264 pKa = 4.05FEE266 pKa = 6.44NDD268 pKa = 2.87MVKK271 pKa = 10.93NNFKK275 pKa = 10.82LNSSEE280 pKa = 4.19GMVHH284 pKa = 7.35SPMLYY289 pKa = 10.27LSLISLFLLIFFSLFLVFSKK309 pKa = 10.39ILKK312 pKa = 9.44PRR314 pKa = 11.84DD315 pKa = 3.15FVSYY319 pKa = 10.58YY320 pKa = 10.59FEE322 pKa = 4.21YY323 pKa = 10.82DD324 pKa = 2.69KK325 pKa = 11.64NRR327 pKa = 11.84IKK329 pKa = 10.51RR330 pKa = 11.84YY331 pKa = 10.1EE332 pKa = 3.83KK333 pKa = 10.43DD334 pKa = 3.13QYY336 pKa = 10.7VV337 pKa = 2.99
MM1 pKa = 7.57KK2 pKa = 10.5GFLAIKK8 pKa = 10.18LSNNEE13 pKa = 4.05YY14 pKa = 10.85YY15 pKa = 9.92PVLNLDD21 pKa = 3.76DD22 pKa = 5.34SSVKK26 pKa = 10.35RR27 pKa = 11.84IILGKK32 pKa = 10.04VAGDD36 pKa = 3.27TSIKK40 pKa = 10.37LNLYY44 pKa = 8.5VSEE47 pKa = 4.84YY48 pKa = 10.96EE49 pKa = 4.78DD50 pKa = 3.89FLDD53 pKa = 4.11PFLVGSFFLDD63 pKa = 3.45NLKK66 pKa = 10.63KK67 pKa = 10.64EE68 pKa = 4.3SLSVNVYY75 pKa = 9.42FKK77 pKa = 11.24VVDD80 pKa = 4.6EE81 pKa = 4.05ILYY84 pKa = 10.73VYY86 pKa = 10.62GEE88 pKa = 4.12SDD90 pKa = 4.62GIQDD94 pKa = 3.58KK95 pKa = 11.51SKK97 pKa = 11.09FDD99 pKa = 3.88LSLVDD104 pKa = 5.2FNQNLKK110 pKa = 10.55GSNRR114 pKa = 11.84GNPLNDD120 pKa = 3.7FKK122 pKa = 11.75SDD124 pKa = 3.17IGSDD128 pKa = 3.49LSVEE132 pKa = 4.18NLGSEE137 pKa = 4.07FDD139 pKa = 4.35LNFEE143 pKa = 4.34EE144 pKa = 6.32KK145 pKa = 10.74DD146 pKa = 3.65DD147 pKa = 4.0FVKK150 pKa = 10.78KK151 pKa = 10.45DD152 pKa = 3.51VSDD155 pKa = 3.77VSMPTINSLDD165 pKa = 3.71LKK167 pKa = 10.13TEE169 pKa = 3.93EE170 pKa = 4.24DD171 pKa = 3.61QKK173 pKa = 11.27IEE175 pKa = 4.3TNVSDD180 pKa = 6.53FEE182 pKa = 5.55DD183 pKa = 4.65FDD185 pKa = 6.05DD186 pKa = 5.67NYY188 pKa = 11.51DD189 pKa = 3.95DD190 pKa = 5.5SLVDD194 pKa = 3.79SKK196 pKa = 11.61KK197 pKa = 11.11LNLEE201 pKa = 4.1NKK203 pKa = 8.42KK204 pKa = 10.32VEE206 pKa = 4.38YY207 pKa = 9.94IDD209 pKa = 4.45EE210 pKa = 4.54KK211 pKa = 11.38SLNLNNFEE219 pKa = 4.04FQKK222 pKa = 10.97LEE224 pKa = 3.94EE225 pKa = 4.42SSLEE229 pKa = 4.05EE230 pKa = 4.19EE231 pKa = 4.68DD232 pKa = 4.73LHH234 pKa = 7.74DD235 pKa = 5.91LSVEE239 pKa = 4.62DD240 pKa = 5.46IIADD244 pKa = 3.4IDD246 pKa = 3.78KK247 pKa = 10.83EE248 pKa = 4.44LIDD251 pKa = 4.04TSDD254 pKa = 3.54EE255 pKa = 4.18DD256 pKa = 3.87VEE258 pKa = 5.07LDD260 pKa = 4.11LRR262 pKa = 11.84EE263 pKa = 4.59DD264 pKa = 4.05FEE266 pKa = 6.44NDD268 pKa = 2.87MVKK271 pKa = 10.93NNFKK275 pKa = 10.82LNSSEE280 pKa = 4.19GMVHH284 pKa = 7.35SPMLYY289 pKa = 10.27LSLISLFLLIFFSLFLVFSKK309 pKa = 10.39ILKK312 pKa = 9.44PRR314 pKa = 11.84DD315 pKa = 3.15FVSYY319 pKa = 10.58YY320 pKa = 10.59FEE322 pKa = 4.21YY323 pKa = 10.82DD324 pKa = 2.69KK325 pKa = 11.64NRR327 pKa = 11.84IKK329 pKa = 10.51RR330 pKa = 11.84YY331 pKa = 10.1EE332 pKa = 3.83KK333 pKa = 10.43DD334 pKa = 3.13QYY336 pKa = 10.7VV337 pKa = 2.99
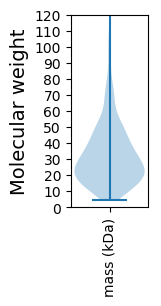
Molecular weight: 39.14 kDa
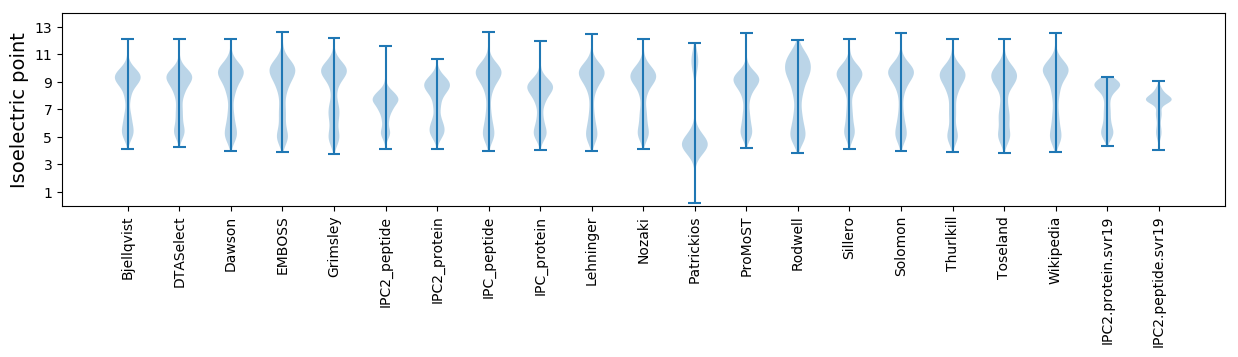
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|O51662|SYT_BORBU Threonine--tRNA ligase OS=Borrelia burgdorferi (strain ATCC 35210 / B31 / CIP 102532 / DSM 4680) OX=224326 GN=thrS PE=3 SV=1
MM1 pKa = 7.67AVPKK5 pKa = 9.82FKK7 pKa = 10.16PSKK10 pKa = 9.35SRR12 pKa = 11.84SRR14 pKa = 11.84TRR16 pKa = 11.84RR17 pKa = 11.84SINMRR22 pKa = 11.84KK23 pKa = 9.83KK24 pKa = 9.1IPQFQEE30 pKa = 3.95CSNCGNLGVRR40 pKa = 11.84HH41 pKa = 6.67RR42 pKa = 11.84ICLKK46 pKa = 9.99CGYY49 pKa = 9.26YY50 pKa = 10.42RR51 pKa = 11.84NNQYY55 pKa = 11.29LEE57 pKa = 4.01IGLL60 pKa = 4.11
MM1 pKa = 7.67AVPKK5 pKa = 9.82FKK7 pKa = 10.16PSKK10 pKa = 9.35SRR12 pKa = 11.84SRR14 pKa = 11.84TRR16 pKa = 11.84RR17 pKa = 11.84SINMRR22 pKa = 11.84KK23 pKa = 9.83KK24 pKa = 9.1IPQFQEE30 pKa = 3.95CSNCGNLGVRR40 pKa = 11.84HH41 pKa = 6.67RR42 pKa = 11.84ICLKK46 pKa = 9.99CGYY49 pKa = 9.26YY50 pKa = 10.42RR51 pKa = 11.84NNQYY55 pKa = 11.29LEE57 pKa = 4.01IGLL60 pKa = 4.11
Molecular weight: 7.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
381647 |
37 |
2166 |
295.9 |
33.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.52 ± 0.065 | 0.744 ± 0.022 |
5.259 ± 0.062 | 7.014 ± 0.075 |
5.937 ± 0.073 | 4.893 ± 0.082 |
1.255 ± 0.022 | 10.294 ± 0.084 |
10.745 ± 0.087 | 10.217 ± 0.076 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.876 ± 0.026 | 7.528 ± 0.078 |
2.431 ± 0.042 | 2.538 ± 0.037 |
3.207 ± 0.044 | 7.41 ± 0.061 |
4.199 ± 0.054 | 5.102 ± 0.065 |
0.473 ± 0.016 | 4.352 ± 0.058 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
