
Koyama Hill virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Orbivirus; Umatilla virus
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
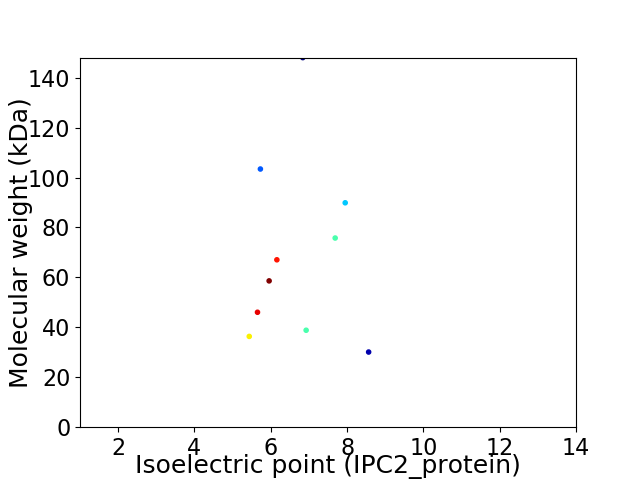
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A077JHF2|A0A077JHF2_9REOV Core protein VP4 OS=Koyama Hill virus OX=1435294 GN=VP4 PE=3 SV=1
MM1 pKa = 7.88RR2 pKa = 11.84IVLLAPGDD10 pKa = 3.72IIKK13 pKa = 10.84NSEE16 pKa = 4.09PEE18 pKa = 4.0LKK20 pKa = 10.19QRR22 pKa = 11.84GIDD25 pKa = 3.51VSVKK29 pKa = 10.56DD30 pKa = 3.82WIEE33 pKa = 3.89NDD35 pKa = 3.4GSQLSEE41 pKa = 4.3GEE43 pKa = 4.23GKK45 pKa = 10.5GGRR48 pKa = 11.84EE49 pKa = 4.25GEE51 pKa = 4.15NGKK54 pKa = 8.91QASANSGRR62 pKa = 11.84RR63 pKa = 11.84DD64 pKa = 3.17GDD66 pKa = 3.42VGRR69 pKa = 11.84DD70 pKa = 3.53GAEE73 pKa = 3.7SSGDD77 pKa = 3.13GKK79 pKa = 11.15ANDD82 pKa = 4.37DD83 pKa = 4.29PRR85 pKa = 11.84PGDD88 pKa = 3.35SGRR91 pKa = 11.84IRR93 pKa = 11.84ADD95 pKa = 3.1GKK97 pKa = 9.57RR98 pKa = 11.84TVEE101 pKa = 4.3TEE103 pKa = 3.93SKK105 pKa = 9.37TGDD108 pKa = 3.11GGEE111 pKa = 3.99IRR113 pKa = 11.84DD114 pKa = 4.18GKK116 pKa = 10.29SASDD120 pKa = 3.59KK121 pKa = 10.58GSKK124 pKa = 8.06NAKK127 pKa = 9.28GNEE130 pKa = 3.7RR131 pKa = 11.84SIDD134 pKa = 3.56STVITSEE141 pKa = 3.99KK142 pKa = 10.0VKK144 pKa = 10.5EE145 pKa = 4.12KK146 pKa = 10.77EE147 pKa = 4.14GTDD150 pKa = 3.49SKK152 pKa = 10.85IAEE155 pKa = 4.06QLAEE159 pKa = 4.1KK160 pKa = 10.57RR161 pKa = 11.84GDD163 pKa = 3.61EE164 pKa = 3.96PSGSNGHH171 pKa = 6.48IYY173 pKa = 10.96VLTRR177 pKa = 11.84AVADD181 pKa = 5.07AINQRR186 pKa = 11.84SGTNLDD192 pKa = 3.47VLKK195 pKa = 10.69SLKK198 pKa = 10.61DD199 pKa = 3.29LGEE202 pKa = 4.19GKK204 pKa = 9.9KK205 pKa = 10.36CYY207 pKa = 9.95QISNGVGKK215 pKa = 10.8LIGVDD220 pKa = 3.02VDD222 pKa = 3.81AVKK225 pKa = 10.49EE226 pKa = 4.0QIDD229 pKa = 3.62ALNEE233 pKa = 3.63LKK235 pKa = 10.7AQYY238 pKa = 10.48DD239 pKa = 3.95GSNVNALLKK248 pKa = 10.0RR249 pKa = 11.84VVRR252 pKa = 11.84IEE254 pKa = 4.07SGGKK258 pKa = 9.33LDD260 pKa = 3.87QVFPKK265 pKa = 10.78GRR267 pKa = 11.84DD268 pKa = 3.47EE269 pKa = 3.95KK270 pKa = 11.07HH271 pKa = 5.65MPGSGVVLATNDD283 pKa = 3.58PKK285 pKa = 11.39YY286 pKa = 10.15IAQANAMFTCPTGDD300 pKa = 3.9TNWKK304 pKa = 8.68EE305 pKa = 4.3VSRR308 pKa = 11.84KK309 pKa = 8.28ATSKK313 pKa = 9.6TSIRR317 pKa = 11.84AYY319 pKa = 9.98SYY321 pKa = 11.53SEE323 pKa = 3.88IGDD326 pKa = 3.41GSEE329 pKa = 3.62IKK331 pKa = 10.43EE332 pKa = 4.27AFLSLIDD339 pKa = 3.84SLL341 pKa = 4.74
MM1 pKa = 7.88RR2 pKa = 11.84IVLLAPGDD10 pKa = 3.72IIKK13 pKa = 10.84NSEE16 pKa = 4.09PEE18 pKa = 4.0LKK20 pKa = 10.19QRR22 pKa = 11.84GIDD25 pKa = 3.51VSVKK29 pKa = 10.56DD30 pKa = 3.82WIEE33 pKa = 3.89NDD35 pKa = 3.4GSQLSEE41 pKa = 4.3GEE43 pKa = 4.23GKK45 pKa = 10.5GGRR48 pKa = 11.84EE49 pKa = 4.25GEE51 pKa = 4.15NGKK54 pKa = 8.91QASANSGRR62 pKa = 11.84RR63 pKa = 11.84DD64 pKa = 3.17GDD66 pKa = 3.42VGRR69 pKa = 11.84DD70 pKa = 3.53GAEE73 pKa = 3.7SSGDD77 pKa = 3.13GKK79 pKa = 11.15ANDD82 pKa = 4.37DD83 pKa = 4.29PRR85 pKa = 11.84PGDD88 pKa = 3.35SGRR91 pKa = 11.84IRR93 pKa = 11.84ADD95 pKa = 3.1GKK97 pKa = 9.57RR98 pKa = 11.84TVEE101 pKa = 4.3TEE103 pKa = 3.93SKK105 pKa = 9.37TGDD108 pKa = 3.11GGEE111 pKa = 3.99IRR113 pKa = 11.84DD114 pKa = 4.18GKK116 pKa = 10.29SASDD120 pKa = 3.59KK121 pKa = 10.58GSKK124 pKa = 8.06NAKK127 pKa = 9.28GNEE130 pKa = 3.7RR131 pKa = 11.84SIDD134 pKa = 3.56STVITSEE141 pKa = 3.99KK142 pKa = 10.0VKK144 pKa = 10.5EE145 pKa = 4.12KK146 pKa = 10.77EE147 pKa = 4.14GTDD150 pKa = 3.49SKK152 pKa = 10.85IAEE155 pKa = 4.06QLAEE159 pKa = 4.1KK160 pKa = 10.57RR161 pKa = 11.84GDD163 pKa = 3.61EE164 pKa = 3.96PSGSNGHH171 pKa = 6.48IYY173 pKa = 10.96VLTRR177 pKa = 11.84AVADD181 pKa = 5.07AINQRR186 pKa = 11.84SGTNLDD192 pKa = 3.47VLKK195 pKa = 10.69SLKK198 pKa = 10.61DD199 pKa = 3.29LGEE202 pKa = 4.19GKK204 pKa = 9.9KK205 pKa = 10.36CYY207 pKa = 9.95QISNGVGKK215 pKa = 10.8LIGVDD220 pKa = 3.02VDD222 pKa = 3.81AVKK225 pKa = 10.49EE226 pKa = 4.0QIDD229 pKa = 3.62ALNEE233 pKa = 3.63LKK235 pKa = 10.7AQYY238 pKa = 10.48DD239 pKa = 3.95GSNVNALLKK248 pKa = 10.0RR249 pKa = 11.84VVRR252 pKa = 11.84IEE254 pKa = 4.07SGGKK258 pKa = 9.33LDD260 pKa = 3.87QVFPKK265 pKa = 10.78GRR267 pKa = 11.84DD268 pKa = 3.47EE269 pKa = 3.95KK270 pKa = 11.07HH271 pKa = 5.65MPGSGVVLATNDD283 pKa = 3.58PKK285 pKa = 11.39YY286 pKa = 10.15IAQANAMFTCPTGDD300 pKa = 3.9TNWKK304 pKa = 8.68EE305 pKa = 4.3VSRR308 pKa = 11.84KK309 pKa = 8.28ATSKK313 pKa = 9.6TSIRR317 pKa = 11.84AYY319 pKa = 9.98SYY321 pKa = 11.53SEE323 pKa = 3.88IGDD326 pKa = 3.41GSEE329 pKa = 3.62IKK331 pKa = 10.43EE332 pKa = 4.27AFLSLIDD339 pKa = 3.84SLL341 pKa = 4.74
Molecular weight: 36.36 kDa
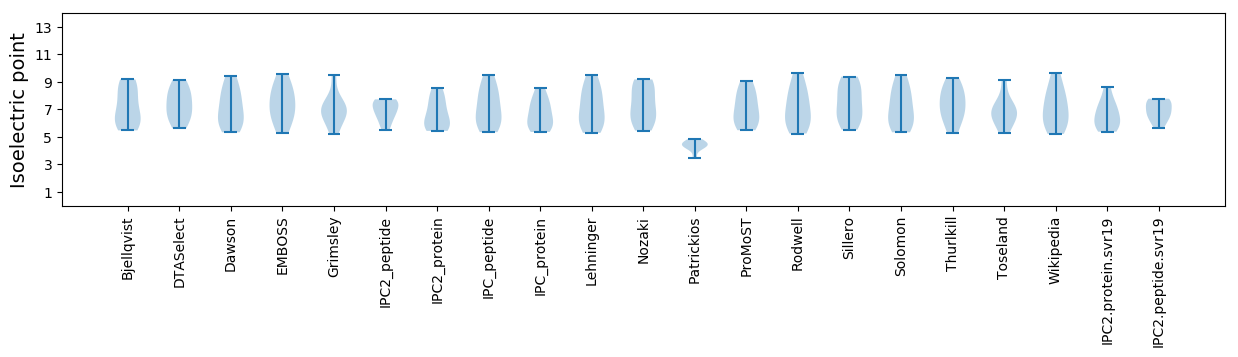
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A077JI40|A0A077JI40_9REOV T2 OS=Koyama Hill virus OX=1435294 GN=VP2 PE=3 SV=1
MM1 pKa = 7.51LEE3 pKa = 3.85AAEE6 pKa = 4.25RR7 pKa = 11.84AQEE10 pKa = 3.98RR11 pKa = 11.84SFTKK15 pKa = 10.56QEE17 pKa = 3.55IQQEE21 pKa = 4.13KK22 pKa = 10.46DD23 pKa = 3.34QIPLIEE29 pKa = 4.45GRR31 pKa = 11.84PNYY34 pKa = 7.62EE35 pKa = 3.82TGQSKK40 pKa = 10.61SSQLKK45 pKa = 8.41PLTRR49 pKa = 11.84FSFLNEE55 pKa = 3.48FNYY58 pKa = 10.18EE59 pKa = 3.98QEE61 pKa = 4.09NGFMYY66 pKa = 10.39QQLQDD71 pKa = 3.31KK72 pKa = 10.05AQALNILTNAVTSTTGANEE91 pKa = 3.81TLKK94 pKa = 11.01NEE96 pKa = 4.11KK97 pKa = 10.21AVFGATSDD105 pKa = 3.76ALKK108 pKa = 10.5DD109 pKa = 3.87DD110 pKa = 4.1PFTRR114 pKa = 11.84YY115 pKa = 7.66TKK117 pKa = 10.14QLAYY121 pKa = 7.81EE122 pKa = 4.2TTIQQINGQMAQKK135 pKa = 9.46RR136 pKa = 11.84RR137 pKa = 11.84KK138 pKa = 8.87MLILDD143 pKa = 4.85LIKK146 pKa = 10.37YY147 pKa = 6.24TLGVILLILLLWNMTVEE164 pKa = 4.69TIDD167 pKa = 3.47EE168 pKa = 4.22EE169 pKa = 4.86SYY171 pKa = 9.09KK172 pKa = 10.45TLLSGLFNRR181 pKa = 11.84NVTDD185 pKa = 3.4VSGMFSAARR194 pKa = 11.84IIRR197 pKa = 11.84RR198 pKa = 11.84YY199 pKa = 8.14TSYY202 pKa = 11.75LEE204 pKa = 4.13MASSIAMTLIIRR216 pKa = 11.84KK217 pKa = 8.82IVQLGMRR224 pKa = 11.84IRR226 pKa = 11.84TLEE229 pKa = 3.66RR230 pKa = 11.84DD231 pKa = 3.33RR232 pKa = 11.84VKK234 pKa = 10.87RR235 pKa = 11.84EE236 pKa = 3.86AYY238 pKa = 8.78NKK240 pKa = 8.0TVARR244 pKa = 11.84VTHH247 pKa = 6.62SGMCPPSAPALEE259 pKa = 4.42STEE262 pKa = 4.06RR263 pKa = 3.48
MM1 pKa = 7.51LEE3 pKa = 3.85AAEE6 pKa = 4.25RR7 pKa = 11.84AQEE10 pKa = 3.98RR11 pKa = 11.84SFTKK15 pKa = 10.56QEE17 pKa = 3.55IQQEE21 pKa = 4.13KK22 pKa = 10.46DD23 pKa = 3.34QIPLIEE29 pKa = 4.45GRR31 pKa = 11.84PNYY34 pKa = 7.62EE35 pKa = 3.82TGQSKK40 pKa = 10.61SSQLKK45 pKa = 8.41PLTRR49 pKa = 11.84FSFLNEE55 pKa = 3.48FNYY58 pKa = 10.18EE59 pKa = 3.98QEE61 pKa = 4.09NGFMYY66 pKa = 10.39QQLQDD71 pKa = 3.31KK72 pKa = 10.05AQALNILTNAVTSTTGANEE91 pKa = 3.81TLKK94 pKa = 11.01NEE96 pKa = 4.11KK97 pKa = 10.21AVFGATSDD105 pKa = 3.76ALKK108 pKa = 10.5DD109 pKa = 3.87DD110 pKa = 4.1PFTRR114 pKa = 11.84YY115 pKa = 7.66TKK117 pKa = 10.14QLAYY121 pKa = 7.81EE122 pKa = 4.2TTIQQINGQMAQKK135 pKa = 9.46RR136 pKa = 11.84RR137 pKa = 11.84KK138 pKa = 8.87MLILDD143 pKa = 4.85LIKK146 pKa = 10.37YY147 pKa = 6.24TLGVILLILLLWNMTVEE164 pKa = 4.69TIDD167 pKa = 3.47EE168 pKa = 4.22EE169 pKa = 4.86SYY171 pKa = 9.09KK172 pKa = 10.45TLLSGLFNRR181 pKa = 11.84NVTDD185 pKa = 3.4VSGMFSAARR194 pKa = 11.84IIRR197 pKa = 11.84RR198 pKa = 11.84YY199 pKa = 8.14TSYY202 pKa = 11.75LEE204 pKa = 4.13MASSIAMTLIIRR216 pKa = 11.84KK217 pKa = 8.82IVQLGMRR224 pKa = 11.84IRR226 pKa = 11.84TLEE229 pKa = 3.66RR230 pKa = 11.84DD231 pKa = 3.33RR232 pKa = 11.84VKK234 pKa = 10.87RR235 pKa = 11.84EE236 pKa = 3.86AYY238 pKa = 8.78NKK240 pKa = 8.0TVARR244 pKa = 11.84VTHH247 pKa = 6.62SGMCPPSAPALEE259 pKa = 4.42STEE262 pKa = 4.06RR263 pKa = 3.48
Molecular weight: 30.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6096 |
263 |
1299 |
609.6 |
69.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.283 ± 0.489 | 1.23 ± 0.209 |
5.676 ± 0.404 | 6.824 ± 0.434 |
3.74 ± 0.358 | 5.807 ± 0.504 |
2.657 ± 0.281 | 6.512 ± 0.281 |
5.299 ± 0.84 | 9.334 ± 0.51 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.887 ± 0.2 | 4.577 ± 0.294 |
3.593 ± 0.395 | 4.085 ± 0.366 |
7.037 ± 0.462 | 6.988 ± 0.589 |
6.217 ± 0.368 | 6.43 ± 0.333 |
1.05 ± 0.165 | 3.757 ± 0.349 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
