
Poaceae-associated gemycircularvirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus poass1; Poaceae associated gemycircularvirus 1
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
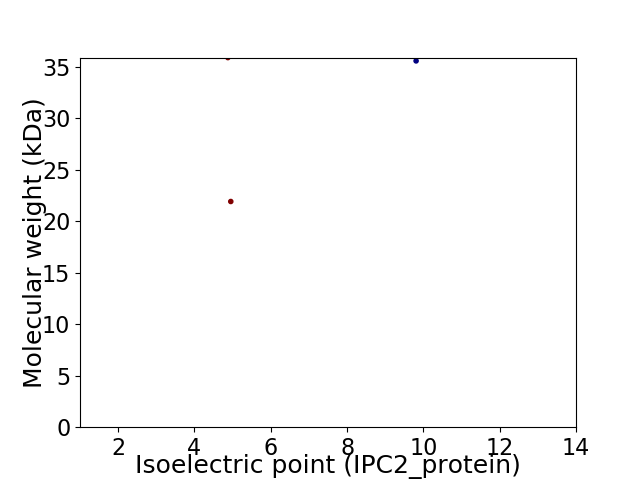
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A0M4G0X9|A0A0M4G0X9_9VIRU Capsid protein OS=Poaceae-associated gemycircularvirus 1 OX=1708497 PE=4 SV=1
MM1 pKa = 6.68THH3 pKa = 7.33FIFSARR9 pKa = 11.84YY10 pKa = 9.01VLLTYY15 pKa = 10.06AQSGDD20 pKa = 3.41LSEE23 pKa = 4.47WDD25 pKa = 3.46ILDD28 pKa = 4.84HH29 pKa = 6.69ISSLGAEE36 pKa = 4.45CIIGRR41 pKa = 11.84EE42 pKa = 4.05DD43 pKa = 3.35HH44 pKa = 7.18ADD46 pKa = 3.64GGTHH50 pKa = 6.1LHH52 pKa = 5.87VFVDD56 pKa = 4.7FGRR59 pKa = 11.84KK60 pKa = 7.88KK61 pKa = 9.87QSRR64 pKa = 11.84RR65 pKa = 11.84SDD67 pKa = 3.46FFDD70 pKa = 3.73VGGHH74 pKa = 6.38HH75 pKa = 7.21PNITPSRR82 pKa = 11.84GRR84 pKa = 11.84PEE86 pKa = 4.18CGYY89 pKa = 11.01DD90 pKa = 3.43YY91 pKa = 10.75AIKK94 pKa = 10.91DD95 pKa = 3.59GDD97 pKa = 4.02VVAGGLARR105 pKa = 11.84PGGGGLPSLADD116 pKa = 3.18KK117 pKa = 10.37WRR119 pKa = 11.84TIVSAEE125 pKa = 3.84SRR127 pKa = 11.84EE128 pKa = 4.04EE129 pKa = 4.15FFDD132 pKa = 4.66LLRR135 pKa = 11.84EE136 pKa = 4.4LDD138 pKa = 3.72PKK140 pKa = 10.54TLVTRR145 pKa = 11.84WSEE148 pKa = 3.74LCRR151 pKa = 11.84YY152 pKa = 9.36ADD154 pKa = 3.21HH155 pKa = 7.31AYY157 pKa = 9.89EE158 pKa = 4.74SKK160 pKa = 10.38PEE162 pKa = 4.2PYY164 pKa = 9.93VGPSGVEE171 pKa = 3.94FEE173 pKa = 4.87LGMVPEE179 pKa = 4.41LVRR182 pKa = 11.84WRR184 pKa = 11.84GDD186 pKa = 3.18TLADD190 pKa = 4.5DD191 pKa = 4.8SEE193 pKa = 5.17LEE195 pKa = 4.18AWAHH199 pKa = 5.65TCIQWEE205 pKa = 4.32SCRR208 pKa = 11.84DD209 pKa = 3.56MFCCGTCRR217 pKa = 11.84EE218 pKa = 3.94VNYY221 pKa = 10.94AVFDD225 pKa = 4.14DD226 pKa = 3.83MRR228 pKa = 11.84GGIGMFPSFKK238 pKa = 10.13EE239 pKa = 3.77WLGAQAVVSVKK250 pKa = 10.32KK251 pKa = 10.67LYY253 pKa = 10.08RR254 pKa = 11.84DD255 pKa = 3.58PVQVPWGRR263 pKa = 11.84PCIWLSNADD272 pKa = 3.65PRR274 pKa = 11.84DD275 pKa = 3.68QIKK278 pKa = 10.81SGLSDD283 pKa = 3.26RR284 pKa = 11.84SSRR287 pKa = 11.84GQIEE291 pKa = 4.58LVEE294 pKa = 4.79NDD296 pKa = 3.65IAWLEE301 pKa = 4.24DD302 pKa = 2.93NCIFVEE308 pKa = 4.05LLEE311 pKa = 4.97PIFRR315 pKa = 11.84ASIGG319 pKa = 3.48
MM1 pKa = 6.68THH3 pKa = 7.33FIFSARR9 pKa = 11.84YY10 pKa = 9.01VLLTYY15 pKa = 10.06AQSGDD20 pKa = 3.41LSEE23 pKa = 4.47WDD25 pKa = 3.46ILDD28 pKa = 4.84HH29 pKa = 6.69ISSLGAEE36 pKa = 4.45CIIGRR41 pKa = 11.84EE42 pKa = 4.05DD43 pKa = 3.35HH44 pKa = 7.18ADD46 pKa = 3.64GGTHH50 pKa = 6.1LHH52 pKa = 5.87VFVDD56 pKa = 4.7FGRR59 pKa = 11.84KK60 pKa = 7.88KK61 pKa = 9.87QSRR64 pKa = 11.84RR65 pKa = 11.84SDD67 pKa = 3.46FFDD70 pKa = 3.73VGGHH74 pKa = 6.38HH75 pKa = 7.21PNITPSRR82 pKa = 11.84GRR84 pKa = 11.84PEE86 pKa = 4.18CGYY89 pKa = 11.01DD90 pKa = 3.43YY91 pKa = 10.75AIKK94 pKa = 10.91DD95 pKa = 3.59GDD97 pKa = 4.02VVAGGLARR105 pKa = 11.84PGGGGLPSLADD116 pKa = 3.18KK117 pKa = 10.37WRR119 pKa = 11.84TIVSAEE125 pKa = 3.84SRR127 pKa = 11.84EE128 pKa = 4.04EE129 pKa = 4.15FFDD132 pKa = 4.66LLRR135 pKa = 11.84EE136 pKa = 4.4LDD138 pKa = 3.72PKK140 pKa = 10.54TLVTRR145 pKa = 11.84WSEE148 pKa = 3.74LCRR151 pKa = 11.84YY152 pKa = 9.36ADD154 pKa = 3.21HH155 pKa = 7.31AYY157 pKa = 9.89EE158 pKa = 4.74SKK160 pKa = 10.38PEE162 pKa = 4.2PYY164 pKa = 9.93VGPSGVEE171 pKa = 3.94FEE173 pKa = 4.87LGMVPEE179 pKa = 4.41LVRR182 pKa = 11.84WRR184 pKa = 11.84GDD186 pKa = 3.18TLADD190 pKa = 4.5DD191 pKa = 4.8SEE193 pKa = 5.17LEE195 pKa = 4.18AWAHH199 pKa = 5.65TCIQWEE205 pKa = 4.32SCRR208 pKa = 11.84DD209 pKa = 3.56MFCCGTCRR217 pKa = 11.84EE218 pKa = 3.94VNYY221 pKa = 10.94AVFDD225 pKa = 4.14DD226 pKa = 3.83MRR228 pKa = 11.84GGIGMFPSFKK238 pKa = 10.13EE239 pKa = 3.77WLGAQAVVSVKK250 pKa = 10.32KK251 pKa = 10.67LYY253 pKa = 10.08RR254 pKa = 11.84DD255 pKa = 3.58PVQVPWGRR263 pKa = 11.84PCIWLSNADD272 pKa = 3.65PRR274 pKa = 11.84DD275 pKa = 3.68QIKK278 pKa = 10.81SGLSDD283 pKa = 3.26RR284 pKa = 11.84SSRR287 pKa = 11.84GQIEE291 pKa = 4.58LVEE294 pKa = 4.79NDD296 pKa = 3.65IAWLEE301 pKa = 4.24DD302 pKa = 2.93NCIFVEE308 pKa = 4.05LLEE311 pKa = 4.97PIFRR315 pKa = 11.84ASIGG319 pKa = 3.48
Molecular weight: 35.92 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0M4G0X9|A0A0M4G0X9_9VIRU Capsid protein OS=Poaceae-associated gemycircularvirus 1 OX=1708497 PE=4 SV=1
MM1 pKa = 7.78AYY3 pKa = 10.13ARR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 7.28STSYY11 pKa = 10.51RR12 pKa = 11.84ARR14 pKa = 11.84KK15 pKa = 8.29PIRR18 pKa = 11.84RR19 pKa = 11.84SVRR22 pKa = 11.84RR23 pKa = 11.84SRR25 pKa = 11.84STVKK29 pKa = 8.65TRR31 pKa = 11.84RR32 pKa = 11.84YY33 pKa = 8.66KK34 pKa = 10.39RR35 pKa = 11.84IYY37 pKa = 8.97RR38 pKa = 11.84RR39 pKa = 11.84PMTKK43 pKa = 9.71RR44 pKa = 11.84RR45 pKa = 11.84ILNATSRR52 pKa = 11.84KK53 pKa = 9.73KK54 pKa = 10.46RR55 pKa = 11.84DD56 pKa = 3.23TMLAASNTNSAGSPLPNTGTRR77 pKa = 11.84AIVYY81 pKa = 9.61GGTSPPTGSSYY92 pKa = 11.61NGGFFLWCATARR104 pKa = 11.84DD105 pKa = 3.93LTINSGPAGAVAQEE119 pKa = 4.18ATRR122 pKa = 11.84TATTCYY128 pKa = 9.73MRR130 pKa = 11.84GLSEE134 pKa = 4.24KK135 pKa = 10.68LRR137 pKa = 11.84VQTNSPAPWFWRR149 pKa = 11.84RR150 pKa = 11.84ICFTIQAAFGVAAPGDD166 pKa = 4.49SPTNTYY172 pKa = 10.9SDD174 pKa = 3.76YY175 pKa = 11.57LEE177 pKa = 4.44NSNGFTRR184 pKa = 11.84LWLNQNINSQGNTTAGITNIVFKK207 pKa = 10.88GAVGRR212 pKa = 11.84DD213 pKa = 2.98WRR215 pKa = 11.84DD216 pKa = 3.43LISAPVDD223 pKa = 3.34TRR225 pKa = 11.84RR226 pKa = 11.84VNLKK230 pKa = 9.49YY231 pKa = 10.82DD232 pKa = 3.19KK233 pKa = 10.87TRR235 pKa = 11.84VFRR238 pKa = 11.84SGNQSGIVRR247 pKa = 11.84EE248 pKa = 4.14LKK250 pKa = 9.94LWHH253 pKa = 6.5PMNRR257 pKa = 11.84NLVYY261 pKa = 10.84DD262 pKa = 4.47DD263 pKa = 5.83DD264 pKa = 4.32EE265 pKa = 4.74VADD268 pKa = 3.87VEE270 pKa = 4.49NTSVYY275 pKa = 10.9SVVDD279 pKa = 3.24KK280 pKa = 11.19RR281 pKa = 11.84GMGNYY286 pKa = 9.16YY287 pKa = 10.43VLDD290 pKa = 4.74IIQPGEE296 pKa = 3.81ASSASDD302 pKa = 4.07LLLIDD307 pKa = 4.31STSSLYY313 pKa = 9.29WHH315 pKa = 7.07EE316 pKa = 4.24KK317 pKa = 9.23
MM1 pKa = 7.78AYY3 pKa = 10.13ARR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 7.28STSYY11 pKa = 10.51RR12 pKa = 11.84ARR14 pKa = 11.84KK15 pKa = 8.29PIRR18 pKa = 11.84RR19 pKa = 11.84SVRR22 pKa = 11.84RR23 pKa = 11.84SRR25 pKa = 11.84STVKK29 pKa = 8.65TRR31 pKa = 11.84RR32 pKa = 11.84YY33 pKa = 8.66KK34 pKa = 10.39RR35 pKa = 11.84IYY37 pKa = 8.97RR38 pKa = 11.84RR39 pKa = 11.84PMTKK43 pKa = 9.71RR44 pKa = 11.84RR45 pKa = 11.84ILNATSRR52 pKa = 11.84KK53 pKa = 9.73KK54 pKa = 10.46RR55 pKa = 11.84DD56 pKa = 3.23TMLAASNTNSAGSPLPNTGTRR77 pKa = 11.84AIVYY81 pKa = 9.61GGTSPPTGSSYY92 pKa = 11.61NGGFFLWCATARR104 pKa = 11.84DD105 pKa = 3.93LTINSGPAGAVAQEE119 pKa = 4.18ATRR122 pKa = 11.84TATTCYY128 pKa = 9.73MRR130 pKa = 11.84GLSEE134 pKa = 4.24KK135 pKa = 10.68LRR137 pKa = 11.84VQTNSPAPWFWRR149 pKa = 11.84RR150 pKa = 11.84ICFTIQAAFGVAAPGDD166 pKa = 4.49SPTNTYY172 pKa = 10.9SDD174 pKa = 3.76YY175 pKa = 11.57LEE177 pKa = 4.44NSNGFTRR184 pKa = 11.84LWLNQNINSQGNTTAGITNIVFKK207 pKa = 10.88GAVGRR212 pKa = 11.84DD213 pKa = 2.98WRR215 pKa = 11.84DD216 pKa = 3.43LISAPVDD223 pKa = 3.34TRR225 pKa = 11.84RR226 pKa = 11.84VNLKK230 pKa = 9.49YY231 pKa = 10.82DD232 pKa = 3.19KK233 pKa = 10.87TRR235 pKa = 11.84VFRR238 pKa = 11.84SGNQSGIVRR247 pKa = 11.84EE248 pKa = 4.14LKK250 pKa = 9.94LWHH253 pKa = 6.5PMNRR257 pKa = 11.84NLVYY261 pKa = 10.84DD262 pKa = 4.47DD263 pKa = 5.83DD264 pKa = 4.32EE265 pKa = 4.74VADD268 pKa = 3.87VEE270 pKa = 4.49NTSVYY275 pKa = 10.9SVVDD279 pKa = 3.24KK280 pKa = 11.19RR281 pKa = 11.84GMGNYY286 pKa = 9.16YY287 pKa = 10.43VLDD290 pKa = 4.74IIQPGEE296 pKa = 3.81ASSASDD302 pKa = 4.07LLLIDD307 pKa = 4.31STSSLYY313 pKa = 9.29WHH315 pKa = 7.07EE316 pKa = 4.24KK317 pKa = 9.23
Molecular weight: 35.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
833 |
197 |
319 |
277.7 |
31.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.963 ± 0.539 | 1.921 ± 0.606 |
7.323 ± 0.981 | 5.642 ± 1.37 |
3.842 ± 0.588 | 8.884 ± 0.854 |
2.281 ± 0.715 | 4.802 ± 0.277 |
3.601 ± 0.355 | 7.683 ± 0.594 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.681 ± 0.092 | 3.241 ± 1.457 |
4.802 ± 0.167 | 1.921 ± 0.253 |
8.884 ± 0.955 | 8.403 ± 0.511 |
5.642 ± 1.601 | 6.242 ± 0.167 |
2.521 ± 0.297 | 3.721 ± 0.503 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
