
Brachypodium distachyon (Purple false brome) (Trachynia distachya)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Stipodae; Brachypodieae; Brachypodium
Average proteome isoelectric point is 6.95
Get precalculated fractions of proteins
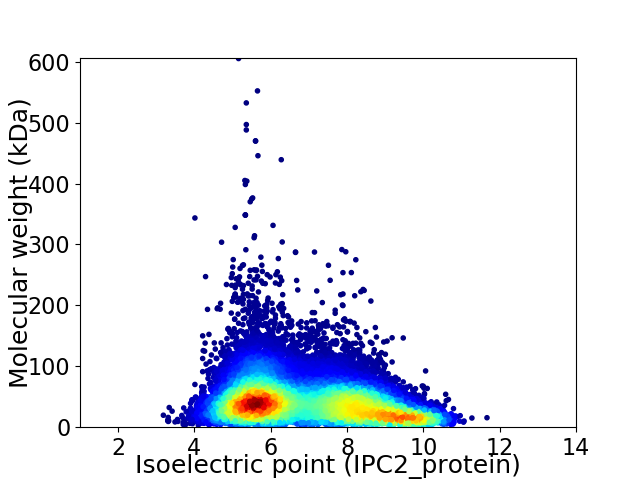
Virtual 2D-PAGE plot for 44786 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I1HEN4|I1HEN4_BRADI Isoform of A0A0Q3IDK8 RanBP2-type domain-containing protein OS=Brachypodium distachyon OX=15368 GN=100836020 PE=3 SV=1
MM1 pKa = 8.05SGMACYY7 pKa = 10.49LLPEE11 pKa = 4.93GGNIFPSDD19 pKa = 3.51EE20 pKa = 4.01EE21 pKa = 4.39LFEE24 pKa = 4.24NQNAEE29 pKa = 4.02QGDD32 pKa = 4.61YY33 pKa = 6.85WTQWNSRR40 pKa = 11.84LSDD43 pKa = 3.96DD44 pKa = 5.23LNTSSIYY51 pKa = 10.11SDD53 pKa = 2.85KK54 pKa = 11.18HH55 pKa = 6.36EE56 pKa = 5.0EE57 pKa = 4.26GSAQCFDD64 pKa = 3.9ADD66 pKa = 3.29EE67 pKa = 4.56HH68 pKa = 6.3QRR70 pKa = 11.84SNQCEE75 pKa = 3.87RR76 pKa = 11.84SSEE79 pKa = 4.06CDD81 pKa = 2.83RR82 pKa = 11.84AASSGCSTGQSEE94 pKa = 4.64EE95 pKa = 4.07QSKK98 pKa = 10.99GPAPLEE104 pKa = 3.84LQQTKK109 pKa = 7.57EE110 pKa = 4.2TNDD113 pKa = 2.9IFLSQFSDD121 pKa = 3.56DD122 pKa = 4.33EE123 pKa = 4.24MRR125 pKa = 11.84MMDD128 pKa = 4.06TPFQALDD135 pKa = 3.47MFPGSMHH142 pKa = 7.23RR143 pKa = 11.84LLSYY147 pKa = 11.23EE148 pKa = 3.94NMLSGVLTDD157 pKa = 3.84STNQVANQDD166 pKa = 3.76RR167 pKa = 11.84NEE169 pKa = 4.2MDD171 pKa = 3.26TMDD174 pKa = 3.44TCGFPLFSHH183 pKa = 7.56DD184 pKa = 5.09LQDD187 pKa = 4.89DD188 pKa = 3.83PSNADD193 pKa = 3.21GSIEE197 pKa = 4.15TLVNPSMDD205 pKa = 3.18KK206 pKa = 11.23SEE208 pKa = 4.0VSTMKK213 pKa = 10.52RR214 pKa = 11.84SWSTADD220 pKa = 3.84IEE222 pKa = 4.65SSSNGEE228 pKa = 3.91GAVLEE233 pKa = 4.16EE234 pKa = 4.68LEE236 pKa = 4.77DD237 pKa = 3.83VVFQVCSPEE246 pKa = 3.58EE247 pKa = 5.13GYY249 pKa = 10.99CFWEE253 pKa = 5.0VYY255 pKa = 10.03FIHH258 pKa = 5.94VTLHH262 pKa = 6.57LYY264 pKa = 9.88MSVSASEE271 pKa = 3.96ADD273 pKa = 4.61KK274 pKa = 11.31EE275 pKa = 4.47DD276 pKa = 3.42ACLPPP281 pKa = 5.54
MM1 pKa = 8.05SGMACYY7 pKa = 10.49LLPEE11 pKa = 4.93GGNIFPSDD19 pKa = 3.51EE20 pKa = 4.01EE21 pKa = 4.39LFEE24 pKa = 4.24NQNAEE29 pKa = 4.02QGDD32 pKa = 4.61YY33 pKa = 6.85WTQWNSRR40 pKa = 11.84LSDD43 pKa = 3.96DD44 pKa = 5.23LNTSSIYY51 pKa = 10.11SDD53 pKa = 2.85KK54 pKa = 11.18HH55 pKa = 6.36EE56 pKa = 5.0EE57 pKa = 4.26GSAQCFDD64 pKa = 3.9ADD66 pKa = 3.29EE67 pKa = 4.56HH68 pKa = 6.3QRR70 pKa = 11.84SNQCEE75 pKa = 3.87RR76 pKa = 11.84SSEE79 pKa = 4.06CDD81 pKa = 2.83RR82 pKa = 11.84AASSGCSTGQSEE94 pKa = 4.64EE95 pKa = 4.07QSKK98 pKa = 10.99GPAPLEE104 pKa = 3.84LQQTKK109 pKa = 7.57EE110 pKa = 4.2TNDD113 pKa = 2.9IFLSQFSDD121 pKa = 3.56DD122 pKa = 4.33EE123 pKa = 4.24MRR125 pKa = 11.84MMDD128 pKa = 4.06TPFQALDD135 pKa = 3.47MFPGSMHH142 pKa = 7.23RR143 pKa = 11.84LLSYY147 pKa = 11.23EE148 pKa = 3.94NMLSGVLTDD157 pKa = 3.84STNQVANQDD166 pKa = 3.76RR167 pKa = 11.84NEE169 pKa = 4.2MDD171 pKa = 3.26TMDD174 pKa = 3.44TCGFPLFSHH183 pKa = 7.56DD184 pKa = 5.09LQDD187 pKa = 4.89DD188 pKa = 3.83PSNADD193 pKa = 3.21GSIEE197 pKa = 4.15TLVNPSMDD205 pKa = 3.18KK206 pKa = 11.23SEE208 pKa = 4.0VSTMKK213 pKa = 10.52RR214 pKa = 11.84SWSTADD220 pKa = 3.84IEE222 pKa = 4.65SSSNGEE228 pKa = 3.91GAVLEE233 pKa = 4.16EE234 pKa = 4.68LEE236 pKa = 4.77DD237 pKa = 3.83VVFQVCSPEE246 pKa = 3.58EE247 pKa = 5.13GYY249 pKa = 10.99CFWEE253 pKa = 5.0VYY255 pKa = 10.03FIHH258 pKa = 5.94VTLHH262 pKa = 6.57LYY264 pKa = 9.88MSVSASEE271 pKa = 3.96ADD273 pKa = 4.61KK274 pKa = 11.31EE275 pKa = 4.47DD276 pKa = 3.42ACLPPP281 pKa = 5.54
Molecular weight: 31.4 kDa
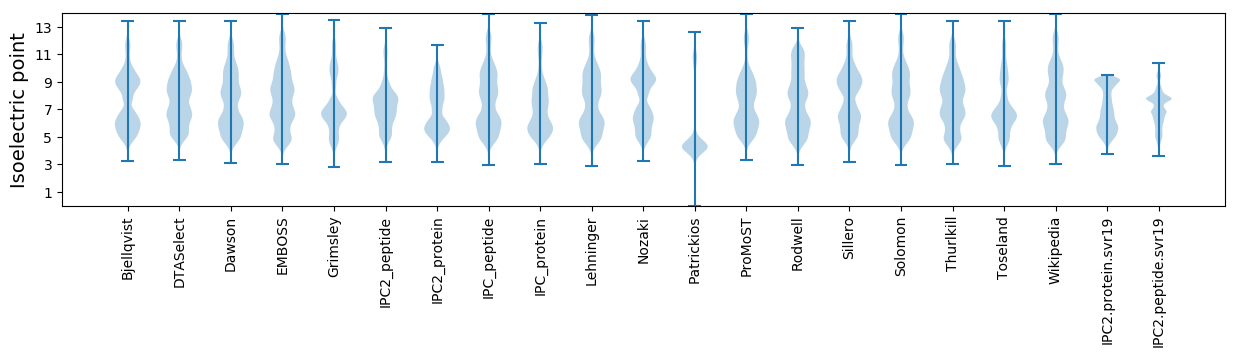
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q3FWT4|A0A0Q3FWT4_BRADI Protein kinase domain-containing protein OS=Brachypodium distachyon OX=15368 GN=100837060 PE=3 SV=1
MM1 pKa = 7.63RR2 pKa = 11.84RR3 pKa = 11.84RR4 pKa = 11.84VRR6 pKa = 11.84PAGARR11 pKa = 11.84TVRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84LRR19 pKa = 11.84AGIRR23 pKa = 11.84TAGARR28 pKa = 11.84TVRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84PRR36 pKa = 11.84AAAGVRR42 pKa = 11.84RR43 pKa = 11.84GPRR46 pKa = 11.84AAAARR51 pKa = 11.84IRR53 pKa = 11.84WRR55 pKa = 11.84PWAAAAAARR64 pKa = 11.84RR65 pKa = 11.84LPSAWRR71 pKa = 11.84LPGAARR77 pKa = 11.84AGRR80 pKa = 11.84LPAGGLPHH88 pKa = 7.1EE89 pKa = 4.89LPRR92 pKa = 11.84AASARR97 pKa = 11.84LHH99 pKa = 6.55GIVPHH104 pKa = 6.0RR105 pKa = 11.84ARR107 pKa = 11.84RR108 pKa = 11.84RR109 pKa = 11.84RR110 pKa = 11.84RR111 pKa = 11.84PWRR114 pKa = 11.84EE115 pKa = 3.32AQGRR119 pKa = 11.84RR120 pKa = 11.84HH121 pKa = 4.52VRR123 pKa = 11.84RR124 pKa = 11.84RR125 pKa = 11.84QARR128 pKa = 11.84PQVEE132 pKa = 4.56VKK134 pKa = 10.69NRR136 pKa = 11.84IATRR140 pKa = 11.84PSVV143 pKa = 3.07
MM1 pKa = 7.63RR2 pKa = 11.84RR3 pKa = 11.84RR4 pKa = 11.84VRR6 pKa = 11.84PAGARR11 pKa = 11.84TVRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84LRR19 pKa = 11.84AGIRR23 pKa = 11.84TAGARR28 pKa = 11.84TVRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84PRR36 pKa = 11.84AAAGVRR42 pKa = 11.84RR43 pKa = 11.84GPRR46 pKa = 11.84AAAARR51 pKa = 11.84IRR53 pKa = 11.84WRR55 pKa = 11.84PWAAAAAARR64 pKa = 11.84RR65 pKa = 11.84LPSAWRR71 pKa = 11.84LPGAARR77 pKa = 11.84AGRR80 pKa = 11.84LPAGGLPHH88 pKa = 7.1EE89 pKa = 4.89LPRR92 pKa = 11.84AASARR97 pKa = 11.84LHH99 pKa = 6.55GIVPHH104 pKa = 6.0RR105 pKa = 11.84ARR107 pKa = 11.84RR108 pKa = 11.84RR109 pKa = 11.84RR110 pKa = 11.84RR111 pKa = 11.84PWRR114 pKa = 11.84EE115 pKa = 3.32AQGRR119 pKa = 11.84RR120 pKa = 11.84HH121 pKa = 4.52VRR123 pKa = 11.84RR124 pKa = 11.84RR125 pKa = 11.84QARR128 pKa = 11.84PQVEE132 pKa = 4.56VKK134 pKa = 10.69NRR136 pKa = 11.84IATRR140 pKa = 11.84PSVV143 pKa = 3.07
Molecular weight: 16.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
18330814 |
23 |
5373 |
409.3 |
45.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.797 ± 0.016 | 1.988 ± 0.006 |
5.389 ± 0.008 | 6.046 ± 0.012 |
3.719 ± 0.006 | 7.127 ± 0.014 |
2.515 ± 0.005 | 4.453 ± 0.008 |
5.037 ± 0.01 | 9.614 ± 0.016 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.433 ± 0.005 | 3.579 ± 0.008 |
5.583 ± 0.014 | 3.564 ± 0.009 |
6.275 ± 0.011 | 8.575 ± 0.014 |
4.816 ± 0.007 | 6.636 ± 0.009 |
1.299 ± 0.004 | 2.554 ± 0.006 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
