
Opuntia virus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Virgaviridae; Tobamovirus; unclassified Tobamovirus
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
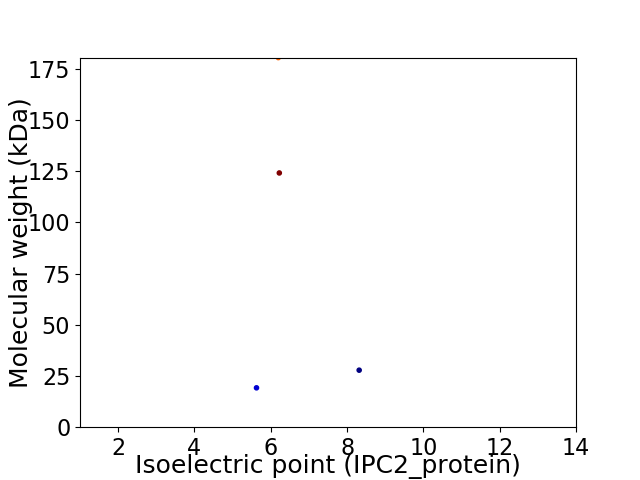
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A2U8U8G9|A0A2U8U8G9_9VIRU Movement protein OS=Opuntia virus 2 OX=2200716 GN=MP PE=3 SV=2
MM1 pKa = 7.87PYY3 pKa = 10.24VGVSTRR9 pKa = 11.84SLALLTPSFVRR20 pKa = 11.84YY21 pKa = 10.08VDD23 pKa = 3.67VLSLIQSILGSQSFQTQSGRR43 pKa = 11.84DD44 pKa = 3.43QLRR47 pKa = 11.84SQLAGLLTAPASRR60 pKa = 11.84DD61 pKa = 3.09VRR63 pKa = 11.84FPEE66 pKa = 3.69EE67 pKa = 4.0TVFVRR72 pKa = 11.84VFDD75 pKa = 4.47SVIEE79 pKa = 3.97PLWLSVLAATDD90 pKa = 3.21TRR92 pKa = 11.84NRR94 pKa = 11.84IIEE97 pKa = 4.12EE98 pKa = 4.01QMVQGPTTPEE108 pKa = 3.48VLNATKK114 pKa = 10.38RR115 pKa = 11.84VDD117 pKa = 3.71DD118 pKa = 3.79ATVACRR124 pKa = 11.84TALSKK129 pKa = 10.71LRR131 pKa = 11.84SGISDD136 pKa = 3.58NLDD139 pKa = 3.34TVCYY143 pKa = 10.5DD144 pKa = 3.2RR145 pKa = 11.84TKK147 pKa = 11.05FEE149 pKa = 4.99LEE151 pKa = 4.68LGSAWLQPGGAPGGGNPAGSGARR174 pKa = 11.84TGTSTT179 pKa = 4.32
MM1 pKa = 7.87PYY3 pKa = 10.24VGVSTRR9 pKa = 11.84SLALLTPSFVRR20 pKa = 11.84YY21 pKa = 10.08VDD23 pKa = 3.67VLSLIQSILGSQSFQTQSGRR43 pKa = 11.84DD44 pKa = 3.43QLRR47 pKa = 11.84SQLAGLLTAPASRR60 pKa = 11.84DD61 pKa = 3.09VRR63 pKa = 11.84FPEE66 pKa = 3.69EE67 pKa = 4.0TVFVRR72 pKa = 11.84VFDD75 pKa = 4.47SVIEE79 pKa = 3.97PLWLSVLAATDD90 pKa = 3.21TRR92 pKa = 11.84NRR94 pKa = 11.84IIEE97 pKa = 4.12EE98 pKa = 4.01QMVQGPTTPEE108 pKa = 3.48VLNATKK114 pKa = 10.38RR115 pKa = 11.84VDD117 pKa = 3.71DD118 pKa = 3.79ATVACRR124 pKa = 11.84TALSKK129 pKa = 10.71LRR131 pKa = 11.84SGISDD136 pKa = 3.58NLDD139 pKa = 3.34TVCYY143 pKa = 10.5DD144 pKa = 3.2RR145 pKa = 11.84TKK147 pKa = 11.05FEE149 pKa = 4.99LEE151 pKa = 4.68LGSAWLQPGGAPGGGNPAGSGARR174 pKa = 11.84TGTSTT179 pKa = 4.32
Molecular weight: 19.23 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2U8U8C4|A0A2U8U8C4_9VIRU Isoform of A0A2U8U8C2 128 kDa hypothetical replicase OS=Opuntia virus 2 OX=2200716 GN=RdRp PE=4 SV=2
MM1 pKa = 7.95AIDD4 pKa = 4.37TYY6 pKa = 11.74SKK8 pKa = 10.76LLEE11 pKa = 4.11PKK13 pKa = 10.34RR14 pKa = 11.84FIKK17 pKa = 10.47PGFWYY22 pKa = 7.55CTPLGKK28 pKa = 10.25SKK30 pKa = 11.18SPLKK34 pKa = 10.43FCNICVTDD42 pKa = 3.59IVKK45 pKa = 10.31VKK47 pKa = 10.67SVGSFSFCLSVYY59 pKa = 9.02KK60 pKa = 9.27TVRR63 pKa = 11.84PDD65 pKa = 2.84HH66 pKa = 7.05RR67 pKa = 11.84SFKK70 pKa = 9.95FARR73 pKa = 11.84LVGVTISGVWHH84 pKa = 6.78FDD86 pKa = 2.72KK87 pKa = 10.91GLPGSAVIALIDD99 pKa = 4.49SRR101 pKa = 11.84ITDD104 pKa = 3.49VNLSILSKK112 pKa = 10.42VVVSASQAGEE122 pKa = 3.92FQFTFSPNFSITMNDD137 pKa = 3.58LKK139 pKa = 10.91RR140 pKa = 11.84NPIEE144 pKa = 5.56LIVDD148 pKa = 4.16LQGLPLCDD156 pKa = 3.32GFEE159 pKa = 4.2PLSLEE164 pKa = 4.26LGFLFYY170 pKa = 11.17YY171 pKa = 10.33SDD173 pKa = 3.84SALSSSLGAKK183 pKa = 9.62IKK185 pKa = 10.51LLQSEE190 pKa = 4.91TNVEE194 pKa = 4.11SGDD197 pKa = 3.8IPDD200 pKa = 3.86HH201 pKa = 6.41VFDD204 pKa = 4.33EE205 pKa = 4.94VVSEE209 pKa = 4.07VDD211 pKa = 3.83LNSVIPKK218 pKa = 9.94ARR220 pKa = 11.84NLRR223 pKa = 11.84NALSVSSKK231 pKa = 10.59SSSALEE237 pKa = 3.74RR238 pKa = 11.84AVVSNKK244 pKa = 8.61FAKK247 pKa = 9.81GAKK250 pKa = 9.07ANKK253 pKa = 9.49RR254 pKa = 3.56
MM1 pKa = 7.95AIDD4 pKa = 4.37TYY6 pKa = 11.74SKK8 pKa = 10.76LLEE11 pKa = 4.11PKK13 pKa = 10.34RR14 pKa = 11.84FIKK17 pKa = 10.47PGFWYY22 pKa = 7.55CTPLGKK28 pKa = 10.25SKK30 pKa = 11.18SPLKK34 pKa = 10.43FCNICVTDD42 pKa = 3.59IVKK45 pKa = 10.31VKK47 pKa = 10.67SVGSFSFCLSVYY59 pKa = 9.02KK60 pKa = 9.27TVRR63 pKa = 11.84PDD65 pKa = 2.84HH66 pKa = 7.05RR67 pKa = 11.84SFKK70 pKa = 9.95FARR73 pKa = 11.84LVGVTISGVWHH84 pKa = 6.78FDD86 pKa = 2.72KK87 pKa = 10.91GLPGSAVIALIDD99 pKa = 4.49SRR101 pKa = 11.84ITDD104 pKa = 3.49VNLSILSKK112 pKa = 10.42VVVSASQAGEE122 pKa = 3.92FQFTFSPNFSITMNDD137 pKa = 3.58LKK139 pKa = 10.91RR140 pKa = 11.84NPIEE144 pKa = 5.56LIVDD148 pKa = 4.16LQGLPLCDD156 pKa = 3.32GFEE159 pKa = 4.2PLSLEE164 pKa = 4.26LGFLFYY170 pKa = 11.17YY171 pKa = 10.33SDD173 pKa = 3.84SALSSSLGAKK183 pKa = 9.62IKK185 pKa = 10.51LLQSEE190 pKa = 4.91TNVEE194 pKa = 4.11SGDD197 pKa = 3.8IPDD200 pKa = 3.86HH201 pKa = 6.41VFDD204 pKa = 4.33EE205 pKa = 4.94VVSEE209 pKa = 4.07VDD211 pKa = 3.83LNSVIPKK218 pKa = 9.94ARR220 pKa = 11.84NLRR223 pKa = 11.84NALSVSSKK231 pKa = 10.59SSSALEE237 pKa = 3.74RR238 pKa = 11.84AVVSNKK244 pKa = 8.61FAKK247 pKa = 9.81GAKK250 pKa = 9.07ANKK253 pKa = 9.49RR254 pKa = 3.56
Molecular weight: 27.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
3131 |
179 |
1598 |
782.8 |
87.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.557 ± 0.361 | 2.236 ± 0.181 |
6.292 ± 0.219 | 5.078 ± 0.239 |
5.717 ± 0.331 | 5.206 ± 0.42 |
2.587 ± 0.492 | 4.919 ± 0.184 |
5.685 ± 0.48 | 9.997 ± 0.263 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.98 ± 0.299 | 4.056 ± 0.241 |
4.503 ± 0.169 | 3.513 ± 0.279 |
5.43 ± 0.281 | 9.358 ± 0.721 |
5.206 ± 0.472 | 8.879 ± 0.375 |
1.054 ± 0.055 | 2.715 ± 0.215 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
