
Carnation cryptic virus 3
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; unclassified Partitiviridae
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
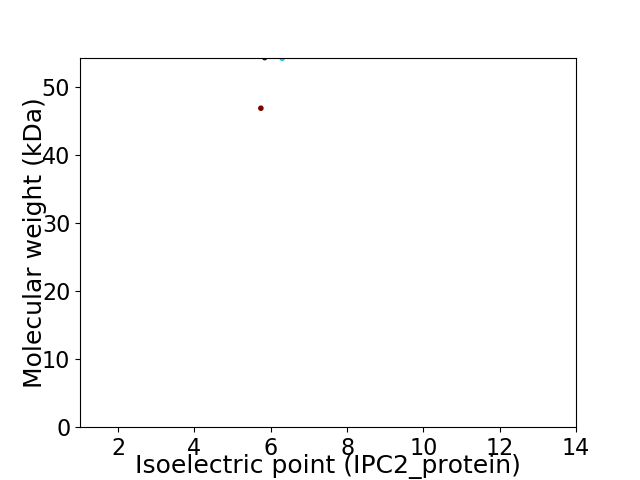
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

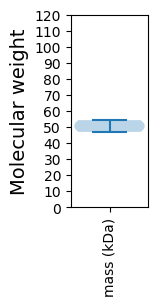
Protein with the lowest isoelectric point:
>tr|A0A1W6C2H2|A0A1W6C2H2_9VIRU Putative RNA-dependent RNA polymerase OS=Carnation cryptic virus 3 OX=1980625 PE=4 SV=1
MM1 pKa = 7.82AEE3 pKa = 3.88NDD5 pKa = 3.5IPAEE9 pKa = 4.6GIPARR14 pKa = 11.84PPPAANPFTGLNRR27 pKa = 11.84DD28 pKa = 3.37EE29 pKa = 5.07LDD31 pKa = 4.32RR32 pKa = 11.84IVEE35 pKa = 4.07QPEE38 pKa = 3.64EE39 pKa = 4.22DD40 pKa = 3.56NQIGLFRR47 pKa = 11.84DD48 pKa = 3.31KK49 pKa = 10.79ARR51 pKa = 11.84SYY53 pKa = 10.86RR54 pKa = 11.84RR55 pKa = 11.84YY56 pKa = 9.72VDD58 pKa = 4.49LRR60 pKa = 11.84TSPLSTRR67 pKa = 11.84LINLYY72 pKa = 10.92SNLFTTHH79 pKa = 5.21WNVFKK84 pKa = 9.99HH85 pKa = 5.02TMSGEE90 pKa = 4.04APAGFASKK98 pKa = 10.55AIYY101 pKa = 9.87LATIYY106 pKa = 10.57VSAWIWDD113 pKa = 3.42LHH115 pKa = 6.48VSIRR119 pKa = 11.84EE120 pKa = 3.98SVRR123 pKa = 11.84RR124 pKa = 11.84ISGTAFNQLFQQDD137 pKa = 4.39AYY139 pKa = 10.24HH140 pKa = 6.09QADD143 pKa = 4.12RR144 pKa = 11.84YY145 pKa = 11.32DD146 pKa = 3.88PFLQHH151 pKa = 7.02LNTIIRR157 pKa = 11.84PTPIHH162 pKa = 6.03QALEE166 pKa = 4.01DD167 pKa = 3.53TLYY170 pKa = 10.68IPVVSRR176 pKa = 11.84NIDD179 pKa = 3.04WANANNNVFGLPSHH193 pKa = 7.02AYY195 pKa = 10.05DD196 pKa = 5.21DD197 pKa = 5.09NILNAILDD205 pKa = 3.88VMDD208 pKa = 4.78DD209 pKa = 5.13RR210 pKa = 11.84RR211 pKa = 11.84NQWSTIPISTNVLGRR226 pKa = 11.84PGWLLDD232 pKa = 3.48YY233 pKa = 10.96RR234 pKa = 11.84LGQAYY239 pKa = 9.88AWFPMEE245 pKa = 4.76ANYY248 pKa = 10.89SQVDD252 pKa = 4.87LIAPHH257 pKa = 7.05ILGTPCTPRR266 pKa = 11.84LGPRR270 pKa = 11.84DD271 pKa = 3.57KK272 pKa = 11.36DD273 pKa = 3.01DD274 pKa = 3.47WQVFRR279 pKa = 11.84YY280 pKa = 9.17NAAPPPGVRR289 pKa = 11.84LNPHH293 pKa = 6.01DD294 pKa = 3.91FRR296 pKa = 11.84RR297 pKa = 11.84ATEE300 pKa = 3.75RR301 pKa = 11.84SFYY304 pKa = 11.11GSAEE308 pKa = 3.79YY309 pKa = 8.48RR310 pKa = 11.84TIEE313 pKa = 4.19HH314 pKa = 6.59AVHH317 pKa = 6.03VFDD320 pKa = 4.28WQATIPPPDD329 pKa = 3.88AATSAASQPGKK340 pKa = 10.24RR341 pKa = 11.84KK342 pKa = 10.05QGDD345 pKa = 3.58SSTSAMALMEE355 pKa = 4.22TDD357 pKa = 4.2KK358 pKa = 11.31DD359 pKa = 3.81PPQPFVPPPPVDD371 pKa = 3.73LNRR374 pKa = 11.84YY375 pKa = 8.91RR376 pKa = 11.84LIDD379 pKa = 3.37WCYY382 pKa = 8.17HH383 pKa = 4.69ARR385 pKa = 11.84VIDD388 pKa = 4.71GEE390 pKa = 4.17DD391 pKa = 3.08QQTRR395 pKa = 11.84NRR397 pKa = 11.84ALRR400 pKa = 11.84NFIFQAGHH408 pKa = 5.57QPTT411 pKa = 3.65
MM1 pKa = 7.82AEE3 pKa = 3.88NDD5 pKa = 3.5IPAEE9 pKa = 4.6GIPARR14 pKa = 11.84PPPAANPFTGLNRR27 pKa = 11.84DD28 pKa = 3.37EE29 pKa = 5.07LDD31 pKa = 4.32RR32 pKa = 11.84IVEE35 pKa = 4.07QPEE38 pKa = 3.64EE39 pKa = 4.22DD40 pKa = 3.56NQIGLFRR47 pKa = 11.84DD48 pKa = 3.31KK49 pKa = 10.79ARR51 pKa = 11.84SYY53 pKa = 10.86RR54 pKa = 11.84RR55 pKa = 11.84YY56 pKa = 9.72VDD58 pKa = 4.49LRR60 pKa = 11.84TSPLSTRR67 pKa = 11.84LINLYY72 pKa = 10.92SNLFTTHH79 pKa = 5.21WNVFKK84 pKa = 9.99HH85 pKa = 5.02TMSGEE90 pKa = 4.04APAGFASKK98 pKa = 10.55AIYY101 pKa = 9.87LATIYY106 pKa = 10.57VSAWIWDD113 pKa = 3.42LHH115 pKa = 6.48VSIRR119 pKa = 11.84EE120 pKa = 3.98SVRR123 pKa = 11.84RR124 pKa = 11.84ISGTAFNQLFQQDD137 pKa = 4.39AYY139 pKa = 10.24HH140 pKa = 6.09QADD143 pKa = 4.12RR144 pKa = 11.84YY145 pKa = 11.32DD146 pKa = 3.88PFLQHH151 pKa = 7.02LNTIIRR157 pKa = 11.84PTPIHH162 pKa = 6.03QALEE166 pKa = 4.01DD167 pKa = 3.53TLYY170 pKa = 10.68IPVVSRR176 pKa = 11.84NIDD179 pKa = 3.04WANANNNVFGLPSHH193 pKa = 7.02AYY195 pKa = 10.05DD196 pKa = 5.21DD197 pKa = 5.09NILNAILDD205 pKa = 3.88VMDD208 pKa = 4.78DD209 pKa = 5.13RR210 pKa = 11.84RR211 pKa = 11.84NQWSTIPISTNVLGRR226 pKa = 11.84PGWLLDD232 pKa = 3.48YY233 pKa = 10.96RR234 pKa = 11.84LGQAYY239 pKa = 9.88AWFPMEE245 pKa = 4.76ANYY248 pKa = 10.89SQVDD252 pKa = 4.87LIAPHH257 pKa = 7.05ILGTPCTPRR266 pKa = 11.84LGPRR270 pKa = 11.84DD271 pKa = 3.57KK272 pKa = 11.36DD273 pKa = 3.01DD274 pKa = 3.47WQVFRR279 pKa = 11.84YY280 pKa = 9.17NAAPPPGVRR289 pKa = 11.84LNPHH293 pKa = 6.01DD294 pKa = 3.91FRR296 pKa = 11.84RR297 pKa = 11.84ATEE300 pKa = 3.75RR301 pKa = 11.84SFYY304 pKa = 11.11GSAEE308 pKa = 3.79YY309 pKa = 8.48RR310 pKa = 11.84TIEE313 pKa = 4.19HH314 pKa = 6.59AVHH317 pKa = 6.03VFDD320 pKa = 4.28WQATIPPPDD329 pKa = 3.88AATSAASQPGKK340 pKa = 10.24RR341 pKa = 11.84KK342 pKa = 10.05QGDD345 pKa = 3.58SSTSAMALMEE355 pKa = 4.22TDD357 pKa = 4.2KK358 pKa = 11.31DD359 pKa = 3.81PPQPFVPPPPVDD371 pKa = 3.73LNRR374 pKa = 11.84YY375 pKa = 8.91RR376 pKa = 11.84LIDD379 pKa = 3.37WCYY382 pKa = 8.17HH383 pKa = 4.69ARR385 pKa = 11.84VIDD388 pKa = 4.71GEE390 pKa = 4.17DD391 pKa = 3.08QQTRR395 pKa = 11.84NRR397 pKa = 11.84ALRR400 pKa = 11.84NFIFQAGHH408 pKa = 5.57QPTT411 pKa = 3.65
Molecular weight: 46.91 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W6C2H2|A0A1W6C2H2_9VIRU Putative RNA-dependent RNA polymerase OS=Carnation cryptic virus 3 OX=1980625 PE=4 SV=1
MM1 pKa = 7.42HH2 pKa = 7.65NINNYY7 pKa = 9.3EE8 pKa = 3.86FTDD11 pKa = 3.85FTEE14 pKa = 4.54EE15 pKa = 3.98VEE17 pKa = 4.44EE18 pKa = 4.19TGRR21 pKa = 11.84THH23 pKa = 5.53HH24 pKa = 6.07HH25 pKa = 4.55VVRR28 pKa = 11.84RR29 pKa = 11.84EE30 pKa = 3.98PEE32 pKa = 3.6VTYY35 pKa = 10.65EE36 pKa = 4.8DD37 pKa = 4.3RR38 pKa = 11.84FASQVLRR45 pKa = 11.84HH46 pKa = 6.47KK47 pKa = 11.28YY48 pKa = 8.74PVLYY52 pKa = 10.61EE53 pKa = 4.13NIISGWSRR61 pKa = 11.84SYY63 pKa = 11.44YY64 pKa = 10.63SGAEE68 pKa = 3.84HH69 pKa = 6.65MKK71 pKa = 10.69SIMQYY76 pKa = 8.11ATPNIKK82 pKa = 10.32LEE84 pKa = 4.04RR85 pKa = 11.84LHH87 pKa = 7.64DD88 pKa = 4.21GVYY91 pKa = 10.43HH92 pKa = 6.61KK93 pKa = 10.89AIANVRR99 pKa = 11.84EE100 pKa = 4.36SLRR103 pKa = 11.84SLPTVRR109 pKa = 11.84AFNVLNEE116 pKa = 4.13LDD118 pKa = 4.06LVPFEE123 pKa = 4.8SSSSAGYY130 pKa = 10.52DD131 pKa = 3.31YY132 pKa = 11.35TGVKK136 pKa = 10.37GPMYY140 pKa = 10.64GEE142 pKa = 3.72NHH144 pKa = 5.37EE145 pKa = 4.21RR146 pKa = 11.84AIRR149 pKa = 11.84RR150 pKa = 11.84AKK152 pKa = 8.59ATLWSAIALDD162 pKa = 4.25GEE164 pKa = 5.11GIDD167 pKa = 3.73HH168 pKa = 7.61VIRR171 pKa = 11.84TYY173 pKa = 11.14VPDD176 pKa = 3.19VGYY179 pKa = 8.69TRR181 pKa = 11.84TQLTDD186 pKa = 3.16LRR188 pKa = 11.84EE189 pKa = 4.0KK190 pKa = 10.37MKK192 pKa = 10.7VRR194 pKa = 11.84GVWGRR199 pKa = 11.84AFHH202 pKa = 6.64YY203 pKa = 10.18ILLEE207 pKa = 4.14GVVAAPLLEE216 pKa = 4.66AFSSADD222 pKa = 3.39TFFHH226 pKa = 6.59IGKK229 pKa = 9.29DD230 pKa = 3.42PTVSVPYY237 pKa = 10.21LLSTIKK243 pKa = 9.62STAEE247 pKa = 3.72WITAIDD253 pKa = 3.74WQSFDD258 pKa = 3.23ATVSRR263 pKa = 11.84FEE265 pKa = 4.11INAAFDD271 pKa = 4.0IIEE274 pKa = 5.38DD275 pKa = 3.41IVSFPNFEE283 pKa = 3.96TEE285 pKa = 3.82QAFEE289 pKa = 3.95ISRR292 pKa = 11.84QLFIHH297 pKa = 6.57KK298 pKa = 9.19MLAAPDD304 pKa = 3.77GKK306 pKa = 10.41IYY308 pKa = 9.8WIHH311 pKa = 6.86KK312 pKa = 7.48GIPSGSYY319 pKa = 7.41FTSTIGSLVNRR330 pKa = 11.84LRR332 pKa = 11.84IEE334 pKa = 4.12YY335 pKa = 8.48MWILKK340 pKa = 9.84FDD342 pKa = 4.63RR343 pKa = 11.84GPKK346 pKa = 8.95ICYY349 pKa = 7.43TQGDD353 pKa = 4.13DD354 pKa = 5.01SLIGDD359 pKa = 4.94DD360 pKa = 4.6EE361 pKa = 5.16LFSPMDD367 pKa = 3.19MTFLVKK373 pKa = 10.17PYY375 pKa = 9.83NWLINVTKK383 pKa = 10.85SMASKK388 pKa = 10.21IPEE391 pKa = 3.85AVTFLGRR398 pKa = 11.84TSIGGLNQRR407 pKa = 11.84DD408 pKa = 4.02LKK410 pKa = 10.89RR411 pKa = 11.84CLRR414 pKa = 11.84LLILPEE420 pKa = 4.1YY421 pKa = 9.34PVTSGDD427 pKa = 2.93ISAYY431 pKa = 8.23RR432 pKa = 11.84AKK434 pKa = 10.66AIWYY438 pKa = 8.85DD439 pKa = 3.33SGLTSEE445 pKa = 4.63VLGEE449 pKa = 3.67IARR452 pKa = 11.84ALRR455 pKa = 11.84RR456 pKa = 11.84KK457 pKa = 10.02YY458 pKa = 10.86GLAEE462 pKa = 3.81KK463 pKa = 10.69SKK465 pKa = 9.88VPKK468 pKa = 10.6YY469 pKa = 10.21LIPWKK474 pKa = 10.7GG475 pKa = 2.94
MM1 pKa = 7.42HH2 pKa = 7.65NINNYY7 pKa = 9.3EE8 pKa = 3.86FTDD11 pKa = 3.85FTEE14 pKa = 4.54EE15 pKa = 3.98VEE17 pKa = 4.44EE18 pKa = 4.19TGRR21 pKa = 11.84THH23 pKa = 5.53HH24 pKa = 6.07HH25 pKa = 4.55VVRR28 pKa = 11.84RR29 pKa = 11.84EE30 pKa = 3.98PEE32 pKa = 3.6VTYY35 pKa = 10.65EE36 pKa = 4.8DD37 pKa = 4.3RR38 pKa = 11.84FASQVLRR45 pKa = 11.84HH46 pKa = 6.47KK47 pKa = 11.28YY48 pKa = 8.74PVLYY52 pKa = 10.61EE53 pKa = 4.13NIISGWSRR61 pKa = 11.84SYY63 pKa = 11.44YY64 pKa = 10.63SGAEE68 pKa = 3.84HH69 pKa = 6.65MKK71 pKa = 10.69SIMQYY76 pKa = 8.11ATPNIKK82 pKa = 10.32LEE84 pKa = 4.04RR85 pKa = 11.84LHH87 pKa = 7.64DD88 pKa = 4.21GVYY91 pKa = 10.43HH92 pKa = 6.61KK93 pKa = 10.89AIANVRR99 pKa = 11.84EE100 pKa = 4.36SLRR103 pKa = 11.84SLPTVRR109 pKa = 11.84AFNVLNEE116 pKa = 4.13LDD118 pKa = 4.06LVPFEE123 pKa = 4.8SSSSAGYY130 pKa = 10.52DD131 pKa = 3.31YY132 pKa = 11.35TGVKK136 pKa = 10.37GPMYY140 pKa = 10.64GEE142 pKa = 3.72NHH144 pKa = 5.37EE145 pKa = 4.21RR146 pKa = 11.84AIRR149 pKa = 11.84RR150 pKa = 11.84AKK152 pKa = 8.59ATLWSAIALDD162 pKa = 4.25GEE164 pKa = 5.11GIDD167 pKa = 3.73HH168 pKa = 7.61VIRR171 pKa = 11.84TYY173 pKa = 11.14VPDD176 pKa = 3.19VGYY179 pKa = 8.69TRR181 pKa = 11.84TQLTDD186 pKa = 3.16LRR188 pKa = 11.84EE189 pKa = 4.0KK190 pKa = 10.37MKK192 pKa = 10.7VRR194 pKa = 11.84GVWGRR199 pKa = 11.84AFHH202 pKa = 6.64YY203 pKa = 10.18ILLEE207 pKa = 4.14GVVAAPLLEE216 pKa = 4.66AFSSADD222 pKa = 3.39TFFHH226 pKa = 6.59IGKK229 pKa = 9.29DD230 pKa = 3.42PTVSVPYY237 pKa = 10.21LLSTIKK243 pKa = 9.62STAEE247 pKa = 3.72WITAIDD253 pKa = 3.74WQSFDD258 pKa = 3.23ATVSRR263 pKa = 11.84FEE265 pKa = 4.11INAAFDD271 pKa = 4.0IIEE274 pKa = 5.38DD275 pKa = 3.41IVSFPNFEE283 pKa = 3.96TEE285 pKa = 3.82QAFEE289 pKa = 3.95ISRR292 pKa = 11.84QLFIHH297 pKa = 6.57KK298 pKa = 9.19MLAAPDD304 pKa = 3.77GKK306 pKa = 10.41IYY308 pKa = 9.8WIHH311 pKa = 6.86KK312 pKa = 7.48GIPSGSYY319 pKa = 7.41FTSTIGSLVNRR330 pKa = 11.84LRR332 pKa = 11.84IEE334 pKa = 4.12YY335 pKa = 8.48MWILKK340 pKa = 9.84FDD342 pKa = 4.63RR343 pKa = 11.84GPKK346 pKa = 8.95ICYY349 pKa = 7.43TQGDD353 pKa = 4.13DD354 pKa = 5.01SLIGDD359 pKa = 4.94DD360 pKa = 4.6EE361 pKa = 5.16LFSPMDD367 pKa = 3.19MTFLVKK373 pKa = 10.17PYY375 pKa = 9.83NWLINVTKK383 pKa = 10.85SMASKK388 pKa = 10.21IPEE391 pKa = 3.85AVTFLGRR398 pKa = 11.84TSIGGLNQRR407 pKa = 11.84DD408 pKa = 4.02LKK410 pKa = 10.89RR411 pKa = 11.84CLRR414 pKa = 11.84LLILPEE420 pKa = 4.1YY421 pKa = 9.34PVTSGDD427 pKa = 2.93ISAYY431 pKa = 8.23RR432 pKa = 11.84AKK434 pKa = 10.66AIWYY438 pKa = 8.85DD439 pKa = 3.33SGLTSEE445 pKa = 4.63VLGEE449 pKa = 3.67IARR452 pKa = 11.84ALRR455 pKa = 11.84RR456 pKa = 11.84KK457 pKa = 10.02YY458 pKa = 10.86GLAEE462 pKa = 3.81KK463 pKa = 10.69SKK465 pKa = 9.88VPKK468 pKa = 10.6YY469 pKa = 10.21LIPWKK474 pKa = 10.7GG475 pKa = 2.94
Molecular weight: 54.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
886 |
411 |
475 |
443.0 |
50.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.126 ± 0.919 | 0.451 ± 0.024 |
6.433 ± 0.912 | 5.418 ± 1.192 |
4.289 ± 0.103 | 5.418 ± 0.7 |
3.047 ± 0.078 | 7.111 ± 0.529 |
3.612 ± 1.287 | 7.901 ± 0.405 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.806 ± 0.233 | 4.402 ± 0.969 |
6.546 ± 1.656 | 3.16 ± 1.15 |
7.336 ± 0.631 | 6.546 ± 0.805 |
5.869 ± 0.184 | 5.53 ± 0.612 |
2.257 ± 0.118 | 4.74 ± 0.407 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
