
Tortoise microvirus 22
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.62
Get precalculated fractions of proteins
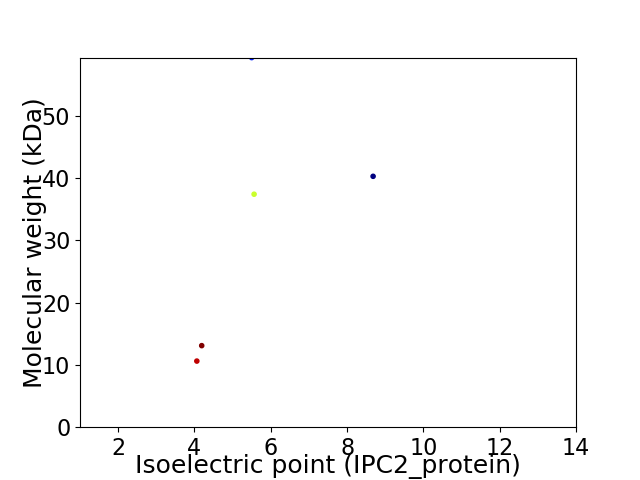
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
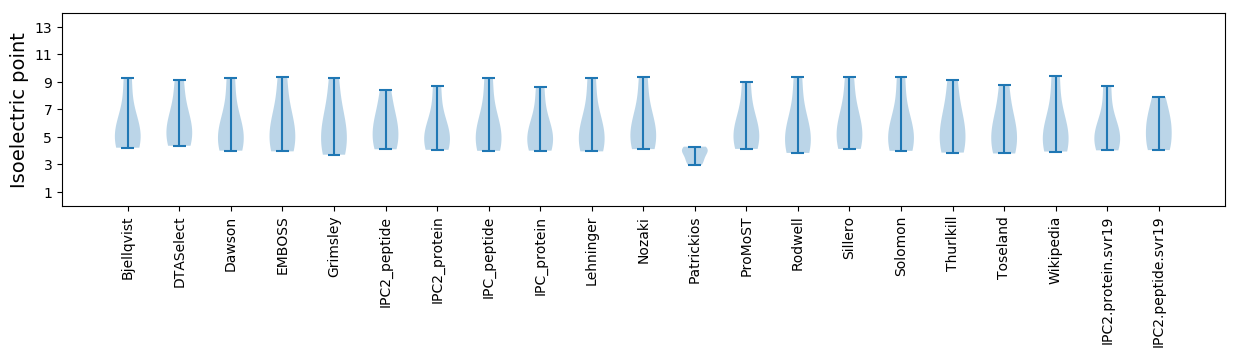
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W6Z2|A0A4P8W6Z2_9VIRU Replication initiation protein OS=Tortoise microvirus 22 OX=2583124 PE=4 SV=1
MM1 pKa = 7.74EE2 pKa = 5.43IVPHH6 pKa = 5.46STVDD10 pKa = 3.58PDD12 pKa = 3.47DD13 pKa = 3.75SFYY16 pKa = 11.67VDD18 pKa = 3.87FFKK21 pKa = 11.11RR22 pKa = 11.84NLITVVCDD30 pKa = 3.35GDD32 pKa = 3.82NADD35 pKa = 4.13SYY37 pKa = 11.87KK38 pKa = 10.73LALGRR43 pKa = 11.84YY44 pKa = 8.62LLFEE48 pKa = 4.25EE49 pKa = 4.94TFSDD53 pKa = 3.93LASAYY58 pKa = 10.22DD59 pKa = 3.89YY60 pKa = 10.67FVSHH64 pKa = 7.1PVPVFLSLSSILRR77 pKa = 11.84EE78 pKa = 3.85IDD80 pKa = 2.92RR81 pKa = 11.84SAASVPEE88 pKa = 4.05SSSDD92 pKa = 3.47VSEE95 pKa = 4.17
MM1 pKa = 7.74EE2 pKa = 5.43IVPHH6 pKa = 5.46STVDD10 pKa = 3.58PDD12 pKa = 3.47DD13 pKa = 3.75SFYY16 pKa = 11.67VDD18 pKa = 3.87FFKK21 pKa = 11.11RR22 pKa = 11.84NLITVVCDD30 pKa = 3.35GDD32 pKa = 3.82NADD35 pKa = 4.13SYY37 pKa = 11.87KK38 pKa = 10.73LALGRR43 pKa = 11.84YY44 pKa = 8.62LLFEE48 pKa = 4.25EE49 pKa = 4.94TFSDD53 pKa = 3.93LASAYY58 pKa = 10.22DD59 pKa = 3.89YY60 pKa = 10.67FVSHH64 pKa = 7.1PVPVFLSLSSILRR77 pKa = 11.84EE78 pKa = 3.85IDD80 pKa = 2.92RR81 pKa = 11.84SAASVPEE88 pKa = 4.05SSSDD92 pKa = 3.47VSEE95 pKa = 4.17
Molecular weight: 10.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W9C1|A0A4P8W9C1_9VIRU Uncharacterized protein OS=Tortoise microvirus 22 OX=2583124 PE=4 SV=1
MM1 pKa = 7.57SCLNPQYY8 pKa = 10.74RR9 pKa = 11.84KK10 pKa = 9.75NPKK13 pKa = 9.11YY14 pKa = 10.45RR15 pKa = 11.84PNKK18 pKa = 9.61SNGGVPPVCPDD29 pKa = 3.2PRR31 pKa = 11.84LMLLRR36 pKa = 11.84YY37 pKa = 10.03DD38 pKa = 3.71CGKK41 pKa = 10.65CYY43 pKa = 9.91EE44 pKa = 4.34CRR46 pKa = 11.84RR47 pKa = 11.84KK48 pKa = 9.81RR49 pKa = 11.84ASHH52 pKa = 4.52WRR54 pKa = 11.84FRR56 pKa = 11.84LLQEE60 pKa = 4.22YY61 pKa = 9.85KK62 pKa = 10.59YY63 pKa = 10.9SPTKK67 pKa = 10.23RR68 pKa = 11.84FHH70 pKa = 6.66FVTFTFSEE78 pKa = 4.49DD79 pKa = 3.47SLFSLRR85 pKa = 11.84SEE87 pKa = 5.33FPDD90 pKa = 5.31LDD92 pKa = 4.28DD93 pKa = 3.87NCIAKK98 pKa = 10.07VAVRR102 pKa = 11.84RR103 pKa = 11.84FLEE106 pKa = 4.58RR107 pKa = 11.84YY108 pKa = 7.01RR109 pKa = 11.84KK110 pKa = 9.81RR111 pKa = 11.84YY112 pKa = 8.95GVSLRR117 pKa = 11.84HH118 pKa = 5.79FLVTEE123 pKa = 4.45LGGKK127 pKa = 9.09NGRR130 pKa = 11.84IHH132 pKa = 6.56LHH134 pKa = 6.36GIVMDD139 pKa = 4.9CKK141 pKa = 9.45AGRR144 pKa = 11.84YY145 pKa = 9.15RR146 pKa = 11.84GSKK149 pKa = 10.2YY150 pKa = 10.43IIDD153 pKa = 3.64MDD155 pKa = 4.37LFHH158 pKa = 7.2SIWRR162 pKa = 11.84YY163 pKa = 9.37GYY165 pKa = 10.96CFFGWCNEE173 pKa = 3.6RR174 pKa = 11.84SISYY178 pKa = 5.81VTKK181 pKa = 10.79YY182 pKa = 10.4IMKK185 pKa = 9.23PDD187 pKa = 4.26PYY189 pKa = 10.62HH190 pKa = 5.22PTFRR194 pKa = 11.84PKK196 pKa = 10.7LLLSPGIGSDD206 pKa = 3.53YY207 pKa = 10.82FRR209 pKa = 11.84DD210 pKa = 3.51LSVLRR215 pKa = 11.84FHH217 pKa = 7.4RR218 pKa = 11.84SSSDD222 pKa = 3.47GVWYY226 pKa = 10.24CVSSTGHH233 pKa = 6.6KK234 pKa = 10.43LSMPRR239 pKa = 11.84YY240 pKa = 8.26YY241 pKa = 10.01RR242 pKa = 11.84DD243 pKa = 3.48KK244 pKa = 10.76IFSEE248 pKa = 4.5DD249 pKa = 2.88EE250 pKa = 4.14RR251 pKa = 11.84LRR253 pKa = 11.84HH254 pKa = 6.75SLDD257 pKa = 4.07LLDD260 pKa = 4.7NPPPFVFRR268 pKa = 11.84GRR270 pKa = 11.84EE271 pKa = 3.96FSSFWSFHH279 pKa = 5.37RR280 pKa = 11.84AVYY283 pKa = 10.57AFYY286 pKa = 10.56DD287 pKa = 3.57YY288 pKa = 10.95TLNLGVSDD296 pKa = 4.5ALCAFRR302 pKa = 11.84DD303 pKa = 3.57NLYY306 pKa = 10.02PDD308 pKa = 3.63EE309 pKa = 5.31SFDD312 pKa = 5.11LFDD315 pKa = 5.08PLIEE319 pKa = 4.04SWSQIRR325 pKa = 11.84IFNDD329 pKa = 3.2YY330 pKa = 10.8LCPQVLGYY338 pKa = 10.82
MM1 pKa = 7.57SCLNPQYY8 pKa = 10.74RR9 pKa = 11.84KK10 pKa = 9.75NPKK13 pKa = 9.11YY14 pKa = 10.45RR15 pKa = 11.84PNKK18 pKa = 9.61SNGGVPPVCPDD29 pKa = 3.2PRR31 pKa = 11.84LMLLRR36 pKa = 11.84YY37 pKa = 10.03DD38 pKa = 3.71CGKK41 pKa = 10.65CYY43 pKa = 9.91EE44 pKa = 4.34CRR46 pKa = 11.84RR47 pKa = 11.84KK48 pKa = 9.81RR49 pKa = 11.84ASHH52 pKa = 4.52WRR54 pKa = 11.84FRR56 pKa = 11.84LLQEE60 pKa = 4.22YY61 pKa = 9.85KK62 pKa = 10.59YY63 pKa = 10.9SPTKK67 pKa = 10.23RR68 pKa = 11.84FHH70 pKa = 6.66FVTFTFSEE78 pKa = 4.49DD79 pKa = 3.47SLFSLRR85 pKa = 11.84SEE87 pKa = 5.33FPDD90 pKa = 5.31LDD92 pKa = 4.28DD93 pKa = 3.87NCIAKK98 pKa = 10.07VAVRR102 pKa = 11.84RR103 pKa = 11.84FLEE106 pKa = 4.58RR107 pKa = 11.84YY108 pKa = 7.01RR109 pKa = 11.84KK110 pKa = 9.81RR111 pKa = 11.84YY112 pKa = 8.95GVSLRR117 pKa = 11.84HH118 pKa = 5.79FLVTEE123 pKa = 4.45LGGKK127 pKa = 9.09NGRR130 pKa = 11.84IHH132 pKa = 6.56LHH134 pKa = 6.36GIVMDD139 pKa = 4.9CKK141 pKa = 9.45AGRR144 pKa = 11.84YY145 pKa = 9.15RR146 pKa = 11.84GSKK149 pKa = 10.2YY150 pKa = 10.43IIDD153 pKa = 3.64MDD155 pKa = 4.37LFHH158 pKa = 7.2SIWRR162 pKa = 11.84YY163 pKa = 9.37GYY165 pKa = 10.96CFFGWCNEE173 pKa = 3.6RR174 pKa = 11.84SISYY178 pKa = 5.81VTKK181 pKa = 10.79YY182 pKa = 10.4IMKK185 pKa = 9.23PDD187 pKa = 4.26PYY189 pKa = 10.62HH190 pKa = 5.22PTFRR194 pKa = 11.84PKK196 pKa = 10.7LLLSPGIGSDD206 pKa = 3.53YY207 pKa = 10.82FRR209 pKa = 11.84DD210 pKa = 3.51LSVLRR215 pKa = 11.84FHH217 pKa = 7.4RR218 pKa = 11.84SSSDD222 pKa = 3.47GVWYY226 pKa = 10.24CVSSTGHH233 pKa = 6.6KK234 pKa = 10.43LSMPRR239 pKa = 11.84YY240 pKa = 8.26YY241 pKa = 10.01RR242 pKa = 11.84DD243 pKa = 3.48KK244 pKa = 10.76IFSEE248 pKa = 4.5DD249 pKa = 2.88EE250 pKa = 4.14RR251 pKa = 11.84LRR253 pKa = 11.84HH254 pKa = 6.75SLDD257 pKa = 4.07LLDD260 pKa = 4.7NPPPFVFRR268 pKa = 11.84GRR270 pKa = 11.84EE271 pKa = 3.96FSSFWSFHH279 pKa = 5.37RR280 pKa = 11.84AVYY283 pKa = 10.57AFYY286 pKa = 10.56DD287 pKa = 3.57YY288 pKa = 10.95TLNLGVSDD296 pKa = 4.5ALCAFRR302 pKa = 11.84DD303 pKa = 3.57NLYY306 pKa = 10.02PDD308 pKa = 3.63EE309 pKa = 5.31SFDD312 pKa = 5.11LFDD315 pKa = 5.08PLIEE319 pKa = 4.04SWSQIRR325 pKa = 11.84IFNDD329 pKa = 3.2YY330 pKa = 10.8LCPQVLGYY338 pKa = 10.82
Molecular weight: 40.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1428 |
95 |
527 |
285.6 |
32.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.073 ± 2.373 | 1.401 ± 0.672 |
7.073 ± 1.068 | 4.342 ± 0.626 |
5.112 ± 1.096 | 5.742 ± 0.741 |
2.381 ± 0.451 | 4.412 ± 0.216 |
3.431 ± 0.435 | 9.034 ± 0.258 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.681 ± 0.195 | 4.412 ± 0.839 |
4.972 ± 1.179 | 4.062 ± 1.544 |
6.513 ± 1.098 | 11.134 ± 1.537 |
4.062 ± 0.551 | 6.793 ± 0.8 |
1.12 ± 0.249 | 5.252 ± 0.806 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
