
Xylella fastidiosa (strain 9a5c)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Xanthomonadaceae; Xylella; Xylella fastidiosa
Average proteome isoelectric point is 7.0
Get precalculated fractions of proteins
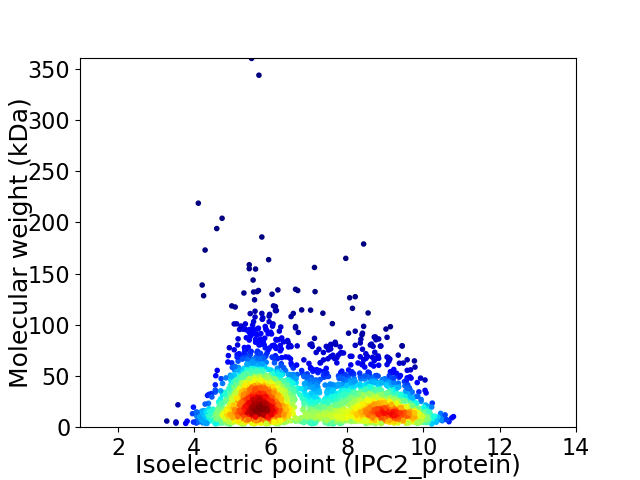
Virtual 2D-PAGE plot for 2772 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9PFC6|Q9PFC6_XYLFA FGE-sulfatase domain-containing protein OS=Xylella fastidiosa (strain 9a5c) OX=160492 GN=XF_0752 PE=4 SV=1
MM1 pKa = 7.79GIALCLRR8 pKa = 11.84HH9 pKa = 7.03ILDD12 pKa = 3.84SDD14 pKa = 3.53ITALAVPSSYY24 pKa = 10.06IAYY27 pKa = 8.93AVPVYY32 pKa = 10.78VVDD35 pKa = 5.06LRR37 pKa = 11.84PDD39 pKa = 3.19TSDD42 pKa = 3.55TNDD45 pKa = 3.07EE46 pKa = 4.17
MM1 pKa = 7.79GIALCLRR8 pKa = 11.84HH9 pKa = 7.03ILDD12 pKa = 3.84SDD14 pKa = 3.53ITALAVPSSYY24 pKa = 10.06IAYY27 pKa = 8.93AVPVYY32 pKa = 10.78VVDD35 pKa = 5.06LRR37 pKa = 11.84PDD39 pKa = 3.19TSDD42 pKa = 3.55TNDD45 pKa = 3.07EE46 pKa = 4.17
Molecular weight: 4.99 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q9PGR0|RS15_XYLFA 30S ribosomal protein S15 OS=Xylella fastidiosa (strain 9a5c) OX=160492 GN=rpsO PE=3 SV=1
MM1 pKa = 7.44SRR3 pKa = 11.84PFSVSEE9 pKa = 4.04TLKK12 pKa = 10.34ADD14 pKa = 3.37CFGRR18 pKa = 11.84ILLVNGKK25 pKa = 8.19GEE27 pKa = 4.01KK28 pKa = 9.95FIRR31 pKa = 11.84RR32 pKa = 11.84DD33 pKa = 3.29LGAVSWWLRR42 pKa = 11.84GLAGWLARR50 pKa = 11.84RR51 pKa = 11.84EE52 pKa = 4.08VAALRR57 pKa = 11.84QLNTLPAVPRR67 pKa = 11.84LLNWDD72 pKa = 4.15GVCLDD77 pKa = 3.98RR78 pKa = 11.84SFLSGQMMYY87 pKa = 10.13QCPPRR92 pKa = 11.84GDD94 pKa = 3.32LAYY97 pKa = 10.59FRR99 pKa = 11.84AACRR103 pKa = 11.84LLQQMHH109 pKa = 6.71RR110 pKa = 11.84CGVVHH115 pKa = 7.2NDD117 pKa = 3.53LAKK120 pKa = 9.53EE121 pKa = 4.09ANWLVLEE128 pKa = 5.1DD129 pKa = 4.48GSPGVIDD136 pKa = 3.71FQLAVRR142 pKa = 11.84GDD144 pKa = 3.78PRR146 pKa = 11.84APWMRR151 pKa = 11.84LLARR155 pKa = 11.84EE156 pKa = 4.35DD157 pKa = 3.42LRR159 pKa = 11.84HH160 pKa = 5.47LLKK163 pKa = 10.4HH164 pKa = 5.85KK165 pKa = 10.61RR166 pKa = 11.84MYY168 pKa = 10.6CPEE171 pKa = 4.15ALTPVEE177 pKa = 4.08RR178 pKa = 11.84RR179 pKa = 11.84LLKK182 pKa = 10.0RR183 pKa = 11.84PSWIRR188 pKa = 11.84RR189 pKa = 11.84LWFATGKK196 pKa = 9.06PVYY199 pKa = 10.02RR200 pKa = 11.84FVTRR204 pKa = 11.84RR205 pKa = 11.84VLHH208 pKa = 6.09WEE210 pKa = 4.07DD211 pKa = 3.46NEE213 pKa = 4.31GRR215 pKa = 11.84GAKK218 pKa = 9.25PP219 pKa = 2.95
MM1 pKa = 7.44SRR3 pKa = 11.84PFSVSEE9 pKa = 4.04TLKK12 pKa = 10.34ADD14 pKa = 3.37CFGRR18 pKa = 11.84ILLVNGKK25 pKa = 8.19GEE27 pKa = 4.01KK28 pKa = 9.95FIRR31 pKa = 11.84RR32 pKa = 11.84DD33 pKa = 3.29LGAVSWWLRR42 pKa = 11.84GLAGWLARR50 pKa = 11.84RR51 pKa = 11.84EE52 pKa = 4.08VAALRR57 pKa = 11.84QLNTLPAVPRR67 pKa = 11.84LLNWDD72 pKa = 4.15GVCLDD77 pKa = 3.98RR78 pKa = 11.84SFLSGQMMYY87 pKa = 10.13QCPPRR92 pKa = 11.84GDD94 pKa = 3.32LAYY97 pKa = 10.59FRR99 pKa = 11.84AACRR103 pKa = 11.84LLQQMHH109 pKa = 6.71RR110 pKa = 11.84CGVVHH115 pKa = 7.2NDD117 pKa = 3.53LAKK120 pKa = 9.53EE121 pKa = 4.09ANWLVLEE128 pKa = 5.1DD129 pKa = 4.48GSPGVIDD136 pKa = 3.71FQLAVRR142 pKa = 11.84GDD144 pKa = 3.78PRR146 pKa = 11.84APWMRR151 pKa = 11.84LLARR155 pKa = 11.84EE156 pKa = 4.35DD157 pKa = 3.42LRR159 pKa = 11.84HH160 pKa = 5.47LLKK163 pKa = 10.4HH164 pKa = 5.85KK165 pKa = 10.61RR166 pKa = 11.84MYY168 pKa = 10.6CPEE171 pKa = 4.15ALTPVEE177 pKa = 4.08RR178 pKa = 11.84RR179 pKa = 11.84LLKK182 pKa = 10.0RR183 pKa = 11.84PSWIRR188 pKa = 11.84RR189 pKa = 11.84LWFATGKK196 pKa = 9.06PVYY199 pKa = 10.02RR200 pKa = 11.84FVTRR204 pKa = 11.84RR205 pKa = 11.84VLHH208 pKa = 6.09WEE210 pKa = 4.07DD211 pKa = 3.46NEE213 pKa = 4.31GRR215 pKa = 11.84GAKK218 pKa = 9.25PP219 pKa = 2.95
Molecular weight: 25.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
743229 |
30 |
3455 |
268.1 |
29.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.235 ± 0.061 | 1.19 ± 0.025 |
5.3 ± 0.039 | 5.071 ± 0.054 |
3.471 ± 0.037 | 7.586 ± 0.063 |
2.743 ± 0.032 | 5.336 ± 0.041 |
3.562 ± 0.043 | 10.714 ± 0.068 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.403 ± 0.028 | 3.358 ± 0.044 |
4.926 ± 0.043 | 4.215 ± 0.035 |
6.694 ± 0.054 | 5.98 ± 0.049 |
5.796 ± 0.085 | 7.336 ± 0.049 |
1.464 ± 0.023 | 2.618 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
