
Aspergillus kawachii (strain NBRC 4308) (White koji mold) (Aspergillus awamori var. kawachi)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; Eurotiomycetes; Eurotiomycetidae; Eurotiales; Aspergillaceae; Aspergillus; Aspergillus luchuensis
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
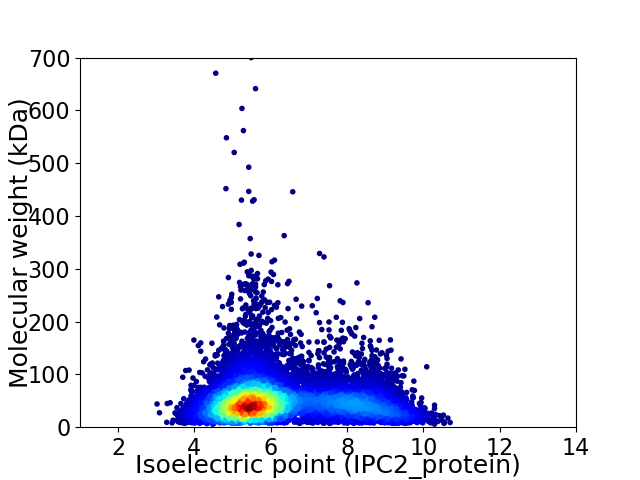
Virtual 2D-PAGE plot for 11491 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|G7XZJ1|G7XZJ1_ASPKW Uncharacterized protein OS=Aspergillus kawachii (strain NBRC 4308) OX=1033177 GN=AKAW_10463 PE=4 SV=1
MM1 pKa = 7.57KK2 pKa = 10.34SYY4 pKa = 10.81GIWSTLATLGSFTLSSAKK22 pKa = 9.29TLNFVAHH29 pKa = 7.12EE30 pKa = 4.56DD31 pKa = 3.72DD32 pKa = 5.65DD33 pKa = 5.68LLFLSPDD40 pKa = 4.47LIRR43 pKa = 11.84DD44 pKa = 3.6ILSGEE49 pKa = 4.39PIRR52 pKa = 11.84TVFLTAGDD60 pKa = 3.85AGMGSDD66 pKa = 3.55YY67 pKa = 10.75WNSRR71 pKa = 11.84EE72 pKa = 4.11DD73 pKa = 3.59GSRR76 pKa = 11.84AAYY79 pKa = 10.58ARR81 pKa = 11.84MAVASNNWDD90 pKa = 3.14EE91 pKa = 4.21DD92 pKa = 4.09VIYY95 pKa = 11.19VGDD98 pKa = 3.94KK99 pKa = 10.37TISLYY104 pKa = 10.02TLQDD108 pKa = 3.76DD109 pKa = 5.22DD110 pKa = 5.76DD111 pKa = 5.1ISLAFMHH118 pKa = 6.84LPDD121 pKa = 4.93GNMDD125 pKa = 3.31GTGFGGTDD133 pKa = 3.76QNDD136 pKa = 3.81SLQKK140 pKa = 10.82LWDD143 pKa = 3.55GDD145 pKa = 3.14IDD147 pKa = 4.19VIRR150 pKa = 11.84TIDD153 pKa = 3.56GSGVQYY159 pKa = 10.69TRR161 pKa = 11.84DD162 pKa = 3.58EE163 pKa = 4.42LLDD166 pKa = 4.16TIAALIDD173 pKa = 4.58DD174 pKa = 5.25FDD176 pKa = 5.85PDD178 pKa = 3.61EE179 pKa = 4.89VKK181 pKa = 10.28TGDD184 pKa = 3.6YY185 pKa = 11.43VNDD188 pKa = 4.35FGDD191 pKa = 4.38GDD193 pKa = 4.1HH194 pKa = 7.19SDD196 pKa = 4.11HH197 pKa = 6.56YY198 pKa = 9.99ATGYY202 pKa = 10.41FVDD205 pKa = 4.5NALQQADD212 pKa = 3.87SDD214 pKa = 4.31ADD216 pKa = 3.51LTGYY220 pKa = 10.02YY221 pKa = 10.2GYY223 pKa = 9.34PIQNMDD229 pKa = 3.52VNLDD233 pKa = 3.65DD234 pKa = 6.3SDD236 pKa = 5.69IEE238 pKa = 4.29DD239 pKa = 3.65KK240 pKa = 11.42TNIFYY245 pKa = 10.61EE246 pKa = 4.16YY247 pKa = 10.63AGYY250 pKa = 8.18DD251 pKa = 3.49TATCATDD258 pKa = 3.84DD259 pKa = 3.99ACSSRR264 pKa = 11.84PEE266 pKa = 4.27SAWLQRR272 pKa = 11.84EE273 pKa = 4.34YY274 pKa = 11.09EE275 pKa = 4.28VV276 pKa = 4.24
MM1 pKa = 7.57KK2 pKa = 10.34SYY4 pKa = 10.81GIWSTLATLGSFTLSSAKK22 pKa = 9.29TLNFVAHH29 pKa = 7.12EE30 pKa = 4.56DD31 pKa = 3.72DD32 pKa = 5.65DD33 pKa = 5.68LLFLSPDD40 pKa = 4.47LIRR43 pKa = 11.84DD44 pKa = 3.6ILSGEE49 pKa = 4.39PIRR52 pKa = 11.84TVFLTAGDD60 pKa = 3.85AGMGSDD66 pKa = 3.55YY67 pKa = 10.75WNSRR71 pKa = 11.84EE72 pKa = 4.11DD73 pKa = 3.59GSRR76 pKa = 11.84AAYY79 pKa = 10.58ARR81 pKa = 11.84MAVASNNWDD90 pKa = 3.14EE91 pKa = 4.21DD92 pKa = 4.09VIYY95 pKa = 11.19VGDD98 pKa = 3.94KK99 pKa = 10.37TISLYY104 pKa = 10.02TLQDD108 pKa = 3.76DD109 pKa = 5.22DD110 pKa = 5.76DD111 pKa = 5.1ISLAFMHH118 pKa = 6.84LPDD121 pKa = 4.93GNMDD125 pKa = 3.31GTGFGGTDD133 pKa = 3.76QNDD136 pKa = 3.81SLQKK140 pKa = 10.82LWDD143 pKa = 3.55GDD145 pKa = 3.14IDD147 pKa = 4.19VIRR150 pKa = 11.84TIDD153 pKa = 3.56GSGVQYY159 pKa = 10.69TRR161 pKa = 11.84DD162 pKa = 3.58EE163 pKa = 4.42LLDD166 pKa = 4.16TIAALIDD173 pKa = 4.58DD174 pKa = 5.25FDD176 pKa = 5.85PDD178 pKa = 3.61EE179 pKa = 4.89VKK181 pKa = 10.28TGDD184 pKa = 3.6YY185 pKa = 11.43VNDD188 pKa = 4.35FGDD191 pKa = 4.38GDD193 pKa = 4.1HH194 pKa = 7.19SDD196 pKa = 4.11HH197 pKa = 6.56YY198 pKa = 9.99ATGYY202 pKa = 10.41FVDD205 pKa = 4.5NALQQADD212 pKa = 3.87SDD214 pKa = 4.31ADD216 pKa = 3.51LTGYY220 pKa = 10.02YY221 pKa = 10.2GYY223 pKa = 9.34PIQNMDD229 pKa = 3.52VNLDD233 pKa = 3.65DD234 pKa = 6.3SDD236 pKa = 5.69IEE238 pKa = 4.29DD239 pKa = 3.65KK240 pKa = 11.42TNIFYY245 pKa = 10.61EE246 pKa = 4.16YY247 pKa = 10.63AGYY250 pKa = 8.18DD251 pKa = 3.49TATCATDD258 pKa = 3.84DD259 pKa = 3.99ACSSRR264 pKa = 11.84PEE266 pKa = 4.27SAWLQRR272 pKa = 11.84EE273 pKa = 4.34YY274 pKa = 11.09EE275 pKa = 4.28VV276 pKa = 4.24
Molecular weight: 30.54 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G7Y2E8|G7Y2E8_ASPKW Zinc knuckle domain protein (Fragment) OS=Aspergillus kawachii (strain NBRC 4308) OX=1033177 GN=AKAW_11469 PE=4 SV=1
MM1 pKa = 7.19FVGSGGAAKK10 pKa = 10.21ISAVCPRR17 pKa = 11.84CWLVSLPISLPISLFLPSPSRR38 pKa = 11.84SPTIFRR44 pKa = 11.84APSVGSLSANTPAHH58 pKa = 6.45WGRR61 pKa = 11.84PRR63 pKa = 11.84SQCRR67 pKa = 11.84VRR69 pKa = 11.84SVLNGCIGRR78 pKa = 11.84EE79 pKa = 3.83THH81 pKa = 5.77VRR83 pKa = 11.84SSKK86 pKa = 9.32TLQEE90 pKa = 3.77SRR92 pKa = 11.84ADD94 pKa = 3.69GRR96 pKa = 11.84SLAMTRR102 pKa = 11.84LSEE105 pKa = 4.08EE106 pKa = 4.25CSWRR110 pKa = 11.84LKK112 pKa = 10.11MGLYY116 pKa = 8.32WWKK119 pKa = 11.03KK120 pKa = 7.53MDD122 pKa = 4.39GPTTPLSARR131 pKa = 11.84EE132 pKa = 4.1ASVSVYY138 pKa = 9.53FVHH141 pKa = 7.31WAPCAHH147 pKa = 6.36SVPTLSQPKK156 pKa = 9.32TGLCGNGIEE165 pKa = 4.33VRR167 pKa = 11.84SRR169 pKa = 11.84VAGAFVCRR177 pKa = 11.84WHH179 pKa = 5.93MSSRR183 pKa = 11.84ASTATDD189 pKa = 3.57LQAPRR194 pKa = 11.84RR195 pKa = 11.84QPEE198 pKa = 4.09LHH200 pKa = 6.71CSTRR204 pKa = 11.84LLLGGCRR211 pKa = 11.84ARR213 pKa = 11.84ASSTGFPAALYY224 pKa = 10.7SDD226 pKa = 4.92ADD228 pKa = 3.43QFTRR232 pKa = 11.84SSAVDD237 pKa = 3.1AHH239 pKa = 5.91TAGAVVFRR247 pKa = 11.84IPRR250 pKa = 11.84PLWHH254 pKa = 6.88IPVPPADD261 pKa = 3.29WLGPGDD267 pKa = 4.11SLVGG271 pKa = 3.57
MM1 pKa = 7.19FVGSGGAAKK10 pKa = 10.21ISAVCPRR17 pKa = 11.84CWLVSLPISLPISLFLPSPSRR38 pKa = 11.84SPTIFRR44 pKa = 11.84APSVGSLSANTPAHH58 pKa = 6.45WGRR61 pKa = 11.84PRR63 pKa = 11.84SQCRR67 pKa = 11.84VRR69 pKa = 11.84SVLNGCIGRR78 pKa = 11.84EE79 pKa = 3.83THH81 pKa = 5.77VRR83 pKa = 11.84SSKK86 pKa = 9.32TLQEE90 pKa = 3.77SRR92 pKa = 11.84ADD94 pKa = 3.69GRR96 pKa = 11.84SLAMTRR102 pKa = 11.84LSEE105 pKa = 4.08EE106 pKa = 4.25CSWRR110 pKa = 11.84LKK112 pKa = 10.11MGLYY116 pKa = 8.32WWKK119 pKa = 11.03KK120 pKa = 7.53MDD122 pKa = 4.39GPTTPLSARR131 pKa = 11.84EE132 pKa = 4.1ASVSVYY138 pKa = 9.53FVHH141 pKa = 7.31WAPCAHH147 pKa = 6.36SVPTLSQPKK156 pKa = 9.32TGLCGNGIEE165 pKa = 4.33VRR167 pKa = 11.84SRR169 pKa = 11.84VAGAFVCRR177 pKa = 11.84WHH179 pKa = 5.93MSSRR183 pKa = 11.84ASTATDD189 pKa = 3.57LQAPRR194 pKa = 11.84RR195 pKa = 11.84QPEE198 pKa = 4.09LHH200 pKa = 6.71CSTRR204 pKa = 11.84LLLGGCRR211 pKa = 11.84ARR213 pKa = 11.84ASSTGFPAALYY224 pKa = 10.7SDD226 pKa = 4.92ADD228 pKa = 3.43QFTRR232 pKa = 11.84SSAVDD237 pKa = 3.1AHH239 pKa = 5.91TAGAVVFRR247 pKa = 11.84IPRR250 pKa = 11.84PLWHH254 pKa = 6.88IPVPPADD261 pKa = 3.29WLGPGDD267 pKa = 4.11SLVGG271 pKa = 3.57
Molecular weight: 29.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5648345 |
48 |
6297 |
491.5 |
54.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.504 ± 0.021 | 1.246 ± 0.009 |
5.673 ± 0.015 | 6.194 ± 0.024 |
3.709 ± 0.014 | 6.791 ± 0.022 |
2.433 ± 0.011 | 4.956 ± 0.015 |
4.458 ± 0.019 | 9.12 ± 0.027 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.181 ± 0.008 | 3.611 ± 0.011 |
6.015 ± 0.028 | 4.041 ± 0.017 |
6.064 ± 0.019 | 8.253 ± 0.025 |
6.063 ± 0.015 | 6.265 ± 0.016 |
1.496 ± 0.008 | 2.926 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
