
Shigella phage pSb-1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Schitoviridae; Enquatrovirinae; Gamaleyavirus; Shigella virus Sb1
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
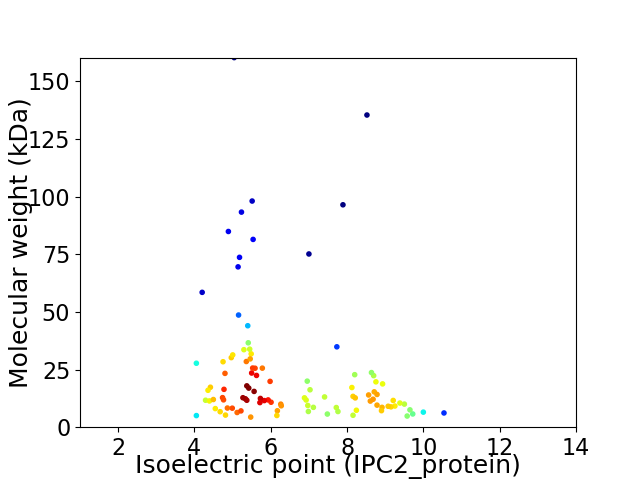
Virtual 2D-PAGE plot for 103 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|V5UPK1|V5UPK1_9CAUD Uncharacterized protein OS=Shigella phage pSb-1 OX=1414738 GN=psb1_0028 PE=4 SV=1
MM1 pKa = 7.23NAHH4 pKa = 6.2TPFDD8 pKa = 4.34AKK10 pKa = 10.72QDD12 pKa = 3.5WTNPYY17 pKa = 9.09CQNSSNDD24 pKa = 3.39PMVDD28 pKa = 3.19ALLGNAYY35 pKa = 9.9HH36 pKa = 6.23VVRR39 pKa = 11.84TVYY42 pKa = 10.84CNLGNLKK49 pKa = 10.53LIYY52 pKa = 10.49DD53 pKa = 4.42FLNKK57 pKa = 10.16YY58 pKa = 10.64GMVLGVQSEE67 pKa = 4.95TEE69 pKa = 4.11LKK71 pKa = 10.53AIPTSASYY79 pKa = 10.84VRR81 pKa = 11.84LYY83 pKa = 11.38GFDD86 pKa = 3.48NTNKK90 pKa = 10.13RR91 pKa = 11.84VVTDD95 pKa = 3.3YY96 pKa = 11.33LYY98 pKa = 11.26VEE100 pKa = 4.84GDD102 pKa = 3.35RR103 pKa = 11.84TGVIPDD109 pKa = 4.44DD110 pKa = 4.34PSATGSWILVATSNSDD126 pKa = 3.07SGGDD130 pKa = 3.64DD131 pKa = 3.9GEE133 pKa = 5.02GKK135 pKa = 10.43ASPPYY140 pKa = 10.19IPYY143 pKa = 10.25SYY145 pKa = 11.28NNGSAIGGEE154 pKa = 4.21TTIPVPAGTVGVPMIVIDD172 pKa = 4.94GYY174 pKa = 9.69TNLVGYY180 pKa = 9.87GFTYY184 pKa = 10.5DD185 pKa = 3.94ASTLTVTLAQPLEE198 pKa = 4.64PGDD201 pKa = 3.99EE202 pKa = 4.0VHH204 pKa = 7.05LFLTGTPAVPDD215 pKa = 3.82NPNVSDD221 pKa = 3.47WVQINWLYY229 pKa = 10.72NGGYY233 pKa = 10.39ASGGEE238 pKa = 3.96QVIAIPYY245 pKa = 7.81TFEE248 pKa = 4.11SVPAIYY254 pKa = 10.52KK255 pKa = 10.29NGEE258 pKa = 3.69RR259 pKa = 11.84YY260 pKa = 9.54YY261 pKa = 11.32AGLADD266 pKa = 4.07KK267 pKa = 10.84SYY269 pKa = 11.59AVDD272 pKa = 3.37AANQRR277 pKa = 11.84ILLTEE282 pKa = 4.13PLATNDD288 pKa = 3.85RR289 pKa = 11.84LIIQIGGEE297 pKa = 4.23STTLIMSDD305 pKa = 3.03RR306 pKa = 11.84TVQEE310 pKa = 4.1VARR313 pKa = 11.84SANVHH318 pKa = 5.93EE319 pKa = 4.7NDD321 pKa = 4.83VILSTNTTQYY331 pKa = 11.07LNGMKK336 pKa = 10.2VIYY339 pKa = 10.22DD340 pKa = 3.41VVGQKK345 pKa = 9.86IYY347 pKa = 11.04GLPTLPTNVYY357 pKa = 9.87INSVSNGQLTYY368 pKa = 10.85SPGNITVDD376 pKa = 4.32LLPMPSEE383 pKa = 4.16ALSALEE389 pKa = 3.83TLEE392 pKa = 4.61NSLNSPLGFTNIGRR406 pKa = 11.84VQSFAALRR414 pKa = 11.84TLVPPSAGARR424 pKa = 11.84VLLSGYY430 pKa = 9.98YY431 pKa = 10.39DD432 pKa = 3.53NSDD435 pKa = 3.32TGGGEE440 pKa = 4.45FIARR444 pKa = 11.84STTGMSEE451 pKa = 4.03VPDD454 pKa = 4.37DD455 pKa = 4.72DD456 pKa = 5.41GGVIATVNSNWYY468 pKa = 8.09WEE470 pKa = 4.24RR471 pKa = 11.84VDD473 pKa = 3.73TNNATVEE480 pKa = 4.09DD481 pKa = 5.26FGAISYY487 pKa = 9.23TGDD490 pKa = 3.61DD491 pKa = 3.5TTGIADD497 pKa = 3.95SSDD500 pKa = 2.64AFQRR504 pKa = 11.84MFNSLNRR511 pKa = 11.84IFSADD516 pKa = 2.96SSKK519 pKa = 10.97RR520 pKa = 11.84FVIDD524 pKa = 3.8APILLKK530 pKa = 10.99VPTLKK535 pKa = 10.77LIYY538 pKa = 9.99QGVVV542 pKa = 2.56
MM1 pKa = 7.23NAHH4 pKa = 6.2TPFDD8 pKa = 4.34AKK10 pKa = 10.72QDD12 pKa = 3.5WTNPYY17 pKa = 9.09CQNSSNDD24 pKa = 3.39PMVDD28 pKa = 3.19ALLGNAYY35 pKa = 9.9HH36 pKa = 6.23VVRR39 pKa = 11.84TVYY42 pKa = 10.84CNLGNLKK49 pKa = 10.53LIYY52 pKa = 10.49DD53 pKa = 4.42FLNKK57 pKa = 10.16YY58 pKa = 10.64GMVLGVQSEE67 pKa = 4.95TEE69 pKa = 4.11LKK71 pKa = 10.53AIPTSASYY79 pKa = 10.84VRR81 pKa = 11.84LYY83 pKa = 11.38GFDD86 pKa = 3.48NTNKK90 pKa = 10.13RR91 pKa = 11.84VVTDD95 pKa = 3.3YY96 pKa = 11.33LYY98 pKa = 11.26VEE100 pKa = 4.84GDD102 pKa = 3.35RR103 pKa = 11.84TGVIPDD109 pKa = 4.44DD110 pKa = 4.34PSATGSWILVATSNSDD126 pKa = 3.07SGGDD130 pKa = 3.64DD131 pKa = 3.9GEE133 pKa = 5.02GKK135 pKa = 10.43ASPPYY140 pKa = 10.19IPYY143 pKa = 10.25SYY145 pKa = 11.28NNGSAIGGEE154 pKa = 4.21TTIPVPAGTVGVPMIVIDD172 pKa = 4.94GYY174 pKa = 9.69TNLVGYY180 pKa = 9.87GFTYY184 pKa = 10.5DD185 pKa = 3.94ASTLTVTLAQPLEE198 pKa = 4.64PGDD201 pKa = 3.99EE202 pKa = 4.0VHH204 pKa = 7.05LFLTGTPAVPDD215 pKa = 3.82NPNVSDD221 pKa = 3.47WVQINWLYY229 pKa = 10.72NGGYY233 pKa = 10.39ASGGEE238 pKa = 3.96QVIAIPYY245 pKa = 7.81TFEE248 pKa = 4.11SVPAIYY254 pKa = 10.52KK255 pKa = 10.29NGEE258 pKa = 3.69RR259 pKa = 11.84YY260 pKa = 9.54YY261 pKa = 11.32AGLADD266 pKa = 4.07KK267 pKa = 10.84SYY269 pKa = 11.59AVDD272 pKa = 3.37AANQRR277 pKa = 11.84ILLTEE282 pKa = 4.13PLATNDD288 pKa = 3.85RR289 pKa = 11.84LIIQIGGEE297 pKa = 4.23STTLIMSDD305 pKa = 3.03RR306 pKa = 11.84TVQEE310 pKa = 4.1VARR313 pKa = 11.84SANVHH318 pKa = 5.93EE319 pKa = 4.7NDD321 pKa = 4.83VILSTNTTQYY331 pKa = 11.07LNGMKK336 pKa = 10.2VIYY339 pKa = 10.22DD340 pKa = 3.41VVGQKK345 pKa = 9.86IYY347 pKa = 11.04GLPTLPTNVYY357 pKa = 9.87INSVSNGQLTYY368 pKa = 10.85SPGNITVDD376 pKa = 4.32LLPMPSEE383 pKa = 4.16ALSALEE389 pKa = 3.83TLEE392 pKa = 4.61NSLNSPLGFTNIGRR406 pKa = 11.84VQSFAALRR414 pKa = 11.84TLVPPSAGARR424 pKa = 11.84VLLSGYY430 pKa = 9.98YY431 pKa = 10.39DD432 pKa = 3.53NSDD435 pKa = 3.32TGGGEE440 pKa = 4.45FIARR444 pKa = 11.84STTGMSEE451 pKa = 4.03VPDD454 pKa = 4.37DD455 pKa = 4.72DD456 pKa = 5.41GGVIATVNSNWYY468 pKa = 8.09WEE470 pKa = 4.24RR471 pKa = 11.84VDD473 pKa = 3.73TNNATVEE480 pKa = 4.09DD481 pKa = 5.26FGAISYY487 pKa = 9.23TGDD490 pKa = 3.61DD491 pKa = 3.5TTGIADD497 pKa = 3.95SSDD500 pKa = 2.64AFQRR504 pKa = 11.84MFNSLNRR511 pKa = 11.84IFSADD516 pKa = 2.96SSKK519 pKa = 10.97RR520 pKa = 11.84FVIDD524 pKa = 3.8APILLKK530 pKa = 10.99VPTLKK535 pKa = 10.77LIYY538 pKa = 9.99QGVVV542 pKa = 2.56
Molecular weight: 58.48 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|V5UQG1|V5UQG1_9CAUD Uncharacterized protein OS=Shigella phage pSb-1 OX=1414738 GN=psb1_0020 PE=4 SV=1
MM1 pKa = 7.17VRR3 pKa = 11.84RR4 pKa = 11.84LKK6 pKa = 10.73RR7 pKa = 11.84RR8 pKa = 11.84VEE10 pKa = 4.15LVPNWKK16 pKa = 9.95RR17 pKa = 11.84CWKK20 pKa = 8.21WASIQISTFGLIMFSAIDD38 pKa = 4.26IIQPMLTGLPRR49 pKa = 11.84HH50 pKa = 6.0ILEE53 pKa = 5.57DD54 pKa = 3.56IPHH57 pKa = 7.03GSNIAIALFALNIVGRR73 pKa = 11.84LIRR76 pKa = 11.84FRR78 pKa = 11.84PKK80 pKa = 10.29EE81 pKa = 4.06DD82 pKa = 3.18SHH84 pKa = 7.49EE85 pKa = 4.23SS86 pKa = 3.3
MM1 pKa = 7.17VRR3 pKa = 11.84RR4 pKa = 11.84LKK6 pKa = 10.73RR7 pKa = 11.84RR8 pKa = 11.84VEE10 pKa = 4.15LVPNWKK16 pKa = 9.95RR17 pKa = 11.84CWKK20 pKa = 8.21WASIQISTFGLIMFSAIDD38 pKa = 4.26IIQPMLTGLPRR49 pKa = 11.84HH50 pKa = 6.0ILEE53 pKa = 5.57DD54 pKa = 3.56IPHH57 pKa = 7.03GSNIAIALFALNIVGRR73 pKa = 11.84LIRR76 pKa = 11.84FRR78 pKa = 11.84PKK80 pKa = 10.29EE81 pKa = 4.06DD82 pKa = 3.18SHH84 pKa = 7.49EE85 pKa = 4.23SS86 pKa = 3.3
Molecular weight: 10.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
21918 |
37 |
1453 |
212.8 |
23.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.491 ± 0.511 | 0.903 ± 0.119 |
5.876 ± 0.139 | 6.314 ± 0.234 |
3.55 ± 0.142 | 6.575 ± 0.238 |
1.752 ± 0.127 | 5.886 ± 0.18 |
6.424 ± 0.281 | 8.258 ± 0.205 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.961 ± 0.114 | 5.717 ± 0.146 |
4.147 ± 0.141 | 4.033 ± 0.245 |
4.581 ± 0.193 | 6.31 ± 0.209 |
6.47 ± 0.251 | 6.912 ± 0.21 |
1.177 ± 0.093 | 3.664 ± 0.198 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
