
Sclerotinia sclerotiorum mycoreovirus 4
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Spinareovirinae; Mycoreovirus; unclassified Mycoreovirus
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
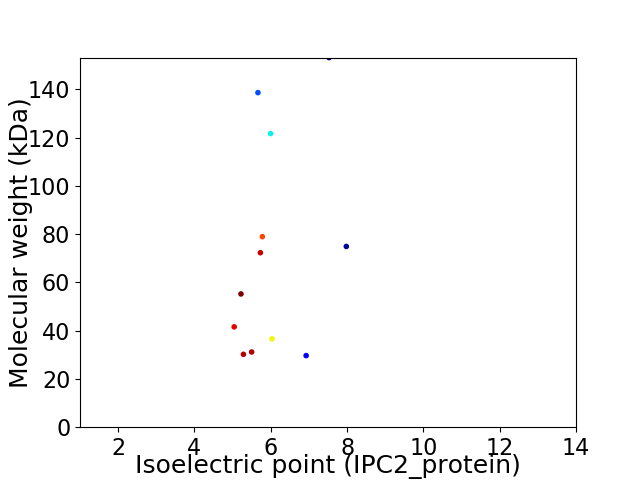
Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A168MTK7|A0A168MTK7_9REOV p10 OS=Sclerotinia sclerotiorum mycoreovirus 4 OX=1840528 PE=4 SV=1
MM1 pKa = 7.45HH2 pKa = 7.97LLTPGLISHH11 pKa = 7.65LSTKK15 pKa = 9.95HH16 pKa = 5.97HH17 pKa = 6.1ATANYY22 pKa = 9.41VPSRR26 pKa = 11.84MAWKK30 pKa = 9.58PYY32 pKa = 7.66QTQLSSNSFVCASAQQLNTASTPQTRR58 pKa = 11.84AQLEE62 pKa = 4.06QRR64 pKa = 11.84VKK66 pKa = 9.39FARR69 pKa = 11.84YY70 pKa = 7.96EE71 pKa = 4.0EE72 pKa = 4.31RR73 pKa = 11.84DD74 pKa = 3.78SIIGYY79 pKa = 10.0ALDD82 pKa = 4.49ASCPLCDD89 pKa = 3.67YY90 pKa = 11.34AEE92 pKa = 4.53SLSDD96 pKa = 3.57TPRR99 pKa = 11.84SPSVGKK105 pKa = 10.29VDD107 pKa = 4.16PSSSSGMGIDD117 pKa = 5.12DD118 pKa = 6.03KK119 pKa = 11.34EE120 pKa = 4.4LSWKK124 pKa = 10.51LDD126 pKa = 3.41LDD128 pKa = 4.35DD129 pKa = 4.28FAKK132 pKa = 10.85AGEE135 pKa = 4.41TVTAAEE141 pKa = 3.84WNEE144 pKa = 4.11RR145 pKa = 11.84FSHH148 pKa = 5.86IPSNFVDD155 pKa = 4.3EE156 pKa = 4.66YY157 pKa = 9.51MLTAASPGIVIVADD171 pKa = 3.78SPAPYY176 pKa = 9.36YY177 pKa = 10.75ASRR180 pKa = 11.84ASKK183 pKa = 10.24FPRR186 pKa = 11.84SRR188 pKa = 11.84QGVIVIPNSINSATSLPNGRR208 pKa = 11.84IIAISTNGHH217 pKa = 5.41EE218 pKa = 4.27ARR220 pKa = 11.84CRR222 pKa = 11.84YY223 pKa = 9.7VDD225 pKa = 3.47LRR227 pKa = 11.84YY228 pKa = 9.64PGRR231 pKa = 11.84KK232 pKa = 8.72RR233 pKa = 11.84LLLEE237 pKa = 4.38VYY239 pKa = 10.72DD240 pKa = 3.88EE241 pKa = 4.58VEE243 pKa = 4.89DD244 pKa = 4.94IEE246 pKa = 5.29EE247 pKa = 4.16EE248 pKa = 4.33DD249 pKa = 3.58PRR251 pKa = 11.84EE252 pKa = 4.25DD253 pKa = 4.74DD254 pKa = 4.51EE255 pKa = 5.57EE256 pKa = 4.03PHH258 pKa = 6.99APLNVMSPILEE269 pKa = 4.01PVIASTSSTSLTGLTLDD286 pKa = 3.83VRR288 pKa = 11.84SASTPPNMPSTSSFSFAEE306 pKa = 4.4DD307 pKa = 3.53PLTNQHH313 pKa = 5.99VPTFDD318 pKa = 3.07HH319 pKa = 6.73RR320 pKa = 11.84RR321 pKa = 11.84AASPLSRR328 pKa = 11.84PDD330 pKa = 3.15TPRR333 pKa = 11.84FMSRR337 pKa = 11.84ARR339 pKa = 11.84CDD341 pKa = 3.12ILLDD345 pKa = 3.87SDD347 pKa = 4.29VSSSMKK353 pKa = 10.51AIASLATSGVITKK366 pKa = 10.46EE367 pKa = 3.97EE368 pKa = 3.89ARR370 pKa = 11.84EE371 pKa = 3.78MLMDD375 pKa = 5.13LGVLL379 pKa = 3.81
MM1 pKa = 7.45HH2 pKa = 7.97LLTPGLISHH11 pKa = 7.65LSTKK15 pKa = 9.95HH16 pKa = 5.97HH17 pKa = 6.1ATANYY22 pKa = 9.41VPSRR26 pKa = 11.84MAWKK30 pKa = 9.58PYY32 pKa = 7.66QTQLSSNSFVCASAQQLNTASTPQTRR58 pKa = 11.84AQLEE62 pKa = 4.06QRR64 pKa = 11.84VKK66 pKa = 9.39FARR69 pKa = 11.84YY70 pKa = 7.96EE71 pKa = 4.0EE72 pKa = 4.31RR73 pKa = 11.84DD74 pKa = 3.78SIIGYY79 pKa = 10.0ALDD82 pKa = 4.49ASCPLCDD89 pKa = 3.67YY90 pKa = 11.34AEE92 pKa = 4.53SLSDD96 pKa = 3.57TPRR99 pKa = 11.84SPSVGKK105 pKa = 10.29VDD107 pKa = 4.16PSSSSGMGIDD117 pKa = 5.12DD118 pKa = 6.03KK119 pKa = 11.34EE120 pKa = 4.4LSWKK124 pKa = 10.51LDD126 pKa = 3.41LDD128 pKa = 4.35DD129 pKa = 4.28FAKK132 pKa = 10.85AGEE135 pKa = 4.41TVTAAEE141 pKa = 3.84WNEE144 pKa = 4.11RR145 pKa = 11.84FSHH148 pKa = 5.86IPSNFVDD155 pKa = 4.3EE156 pKa = 4.66YY157 pKa = 9.51MLTAASPGIVIVADD171 pKa = 3.78SPAPYY176 pKa = 9.36YY177 pKa = 10.75ASRR180 pKa = 11.84ASKK183 pKa = 10.24FPRR186 pKa = 11.84SRR188 pKa = 11.84QGVIVIPNSINSATSLPNGRR208 pKa = 11.84IIAISTNGHH217 pKa = 5.41EE218 pKa = 4.27ARR220 pKa = 11.84CRR222 pKa = 11.84YY223 pKa = 9.7VDD225 pKa = 3.47LRR227 pKa = 11.84YY228 pKa = 9.64PGRR231 pKa = 11.84KK232 pKa = 8.72RR233 pKa = 11.84LLLEE237 pKa = 4.38VYY239 pKa = 10.72DD240 pKa = 3.88EE241 pKa = 4.58VEE243 pKa = 4.89DD244 pKa = 4.94IEE246 pKa = 5.29EE247 pKa = 4.16EE248 pKa = 4.33DD249 pKa = 3.58PRR251 pKa = 11.84EE252 pKa = 4.25DD253 pKa = 4.74DD254 pKa = 4.51EE255 pKa = 5.57EE256 pKa = 4.03PHH258 pKa = 6.99APLNVMSPILEE269 pKa = 4.01PVIASTSSTSLTGLTLDD286 pKa = 3.83VRR288 pKa = 11.84SASTPPNMPSTSSFSFAEE306 pKa = 4.4DD307 pKa = 3.53PLTNQHH313 pKa = 5.99VPTFDD318 pKa = 3.07HH319 pKa = 6.73RR320 pKa = 11.84RR321 pKa = 11.84AASPLSRR328 pKa = 11.84PDD330 pKa = 3.15TPRR333 pKa = 11.84FMSRR337 pKa = 11.84ARR339 pKa = 11.84CDD341 pKa = 3.12ILLDD345 pKa = 3.87SDD347 pKa = 4.29VSSSMKK353 pKa = 10.51AIASLATSGVITKK366 pKa = 10.46EE367 pKa = 3.97EE368 pKa = 3.89ARR370 pKa = 11.84EE371 pKa = 3.78MLMDD375 pKa = 5.13LGVLL379 pKa = 3.81
Molecular weight: 41.57 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A168MTI0|A0A168MTI0_9REOV p7 OS=Sclerotinia sclerotiorum mycoreovirus 4 OX=1840528 PE=4 SV=1
MM1 pKa = 7.38NALADD6 pKa = 4.35PNSSNGPPLSDD17 pKa = 3.03VTFPLIRR24 pKa = 11.84RR25 pKa = 11.84SFSHH29 pKa = 5.92ATYY32 pKa = 7.61TTLPLPTDD40 pKa = 3.7IYY42 pKa = 11.49QMLRR46 pKa = 11.84CVEE49 pKa = 4.28VYY51 pKa = 10.66LEE53 pKa = 4.01RR54 pKa = 11.84LYY56 pKa = 10.69TADD59 pKa = 2.84VTRR62 pKa = 11.84YY63 pKa = 9.19VDD65 pKa = 3.37VVSNYY70 pKa = 9.92RR71 pKa = 11.84KK72 pKa = 8.01TSVARR77 pKa = 11.84AVPHH81 pKa = 5.96IFVVNSMEE89 pKa = 4.2KK90 pKa = 10.58FSTSYY95 pKa = 9.62STLLSKK101 pKa = 10.82SPHH104 pKa = 7.16DD105 pKa = 4.0YY106 pKa = 10.54VSSDD110 pKa = 3.22VTVVSNPTSQDD121 pKa = 2.71IIRR124 pKa = 11.84SIRR127 pKa = 11.84AHH129 pKa = 5.65KK130 pKa = 10.22HH131 pKa = 3.51VVCNVVPIGDD141 pKa = 3.77FSDD144 pKa = 5.02AILMPNASISDD155 pKa = 3.7CVKK158 pKa = 8.48YY159 pKa = 10.76LRR161 pKa = 11.84RR162 pKa = 11.84NFHH165 pKa = 6.14ALLRR169 pKa = 11.84RR170 pKa = 11.84LSFGHH175 pKa = 6.37GCYY178 pKa = 10.1SCSNVLQLSLPPWLSVLASGWQRR201 pKa = 11.84FKK203 pKa = 10.66PTGSDD208 pKa = 2.63TDD210 pKa = 3.95FAQFYY215 pKa = 7.28LTPARR220 pKa = 11.84PQLSVINSISDD231 pKa = 3.42LSDD234 pKa = 4.07FIINRR239 pKa = 11.84LQANVDD245 pKa = 3.3VVYY248 pKa = 10.69FDD250 pKa = 6.15DD251 pKa = 4.97VLTTDD256 pKa = 3.43GRR258 pKa = 11.84TIAEE262 pKa = 4.63KK263 pKa = 10.39IRR265 pKa = 11.84ATGAQFNVARR275 pKa = 11.84DD276 pKa = 3.61MTHH279 pKa = 6.76FNIIKK284 pKa = 9.62QRR286 pKa = 11.84YY287 pKa = 8.28LEE289 pKa = 3.94QTSIRR294 pKa = 11.84RR295 pKa = 11.84RR296 pKa = 11.84RR297 pKa = 11.84QHH299 pKa = 6.42KK300 pKa = 10.37KK301 pKa = 9.19EE302 pKa = 3.71LHH304 pKa = 5.72ASDD307 pKa = 3.95RR308 pKa = 11.84KK309 pKa = 9.36QNIVARR315 pKa = 11.84VFPPVYY321 pKa = 10.33LVDD324 pKa = 4.47LSVSIRR330 pKa = 11.84AWNSAMPTQQQRR342 pKa = 11.84RR343 pKa = 11.84LSRR346 pKa = 11.84VYY348 pKa = 10.87SDD350 pKa = 4.68LVNLSKK356 pKa = 10.93RR357 pKa = 11.84PLTVLHH363 pKa = 6.44MIAWNGPSVVSTDD376 pKa = 2.91SDD378 pKa = 3.43VWQDD382 pKa = 3.23FTEE385 pKa = 3.98WLFCVVQNVDD395 pKa = 2.79EE396 pKa = 4.5SMARR400 pKa = 11.84LLTAIPYY407 pKa = 8.59MSVEE411 pKa = 4.13IKK413 pKa = 10.49ISGSGFGTWIDD424 pKa = 3.5LRR426 pKa = 11.84NWTEE430 pKa = 4.05GFSLSKK436 pKa = 10.41ILVGRR441 pKa = 11.84TYY443 pKa = 10.47MVGKK447 pKa = 9.93KK448 pKa = 10.13NSGKK452 pKa = 10.94GMIGKK457 pKa = 8.64LIWRR461 pKa = 11.84LGVPVVDD468 pKa = 3.5SDD470 pKa = 4.06DD471 pKa = 3.7YY472 pKa = 11.99GRR474 pKa = 11.84VLLIAEE480 pKa = 4.21NTGVSLEE487 pKa = 4.19DD488 pKa = 3.29AVRR491 pKa = 11.84CHH493 pKa = 6.59FSRR496 pKa = 11.84SYY498 pKa = 11.52ADD500 pKa = 4.03RR501 pKa = 11.84DD502 pKa = 3.7CAPTLFEE509 pKa = 5.11TIMDD513 pKa = 5.3DD514 pKa = 2.82IVTRR518 pKa = 11.84LSIKK522 pKa = 9.96RR523 pKa = 11.84YY524 pKa = 9.39SIPEE528 pKa = 3.78VDD530 pKa = 4.58HH531 pKa = 6.89PALMMFGHH539 pKa = 7.57AYY541 pKa = 10.45DD542 pKa = 4.53EE543 pKa = 4.18LTKK546 pKa = 10.25KK547 pKa = 10.55YY548 pKa = 10.37KK549 pKa = 10.37YY550 pKa = 10.74ADD552 pKa = 3.64FEE554 pKa = 5.14RR555 pKa = 11.84MVRR558 pKa = 11.84SFISHH563 pKa = 6.5HH564 pKa = 5.83GVRR567 pKa = 11.84APDD570 pKa = 3.68GSLLKK575 pKa = 10.59VDD577 pKa = 4.08DD578 pKa = 4.2RR579 pKa = 11.84FVFSTHH585 pKa = 6.2CSEE588 pKa = 5.02EE589 pKa = 3.95ATQVLGANYY598 pKa = 8.8MFQLSTAIDD607 pKa = 3.85SYY609 pKa = 11.72VGVLLRR615 pKa = 11.84GQHH618 pKa = 6.52DD619 pKa = 3.83NSISEE624 pKa = 4.46LMLAVYY630 pKa = 10.0YY631 pKa = 10.69DD632 pKa = 4.57RR633 pKa = 11.84IHH635 pKa = 7.83VNIFDD640 pKa = 4.57LVPTGVVCTVLRR652 pKa = 11.84TGLARR657 pKa = 11.84LVPKK661 pKa = 10.64AA662 pKa = 3.7
MM1 pKa = 7.38NALADD6 pKa = 4.35PNSSNGPPLSDD17 pKa = 3.03VTFPLIRR24 pKa = 11.84RR25 pKa = 11.84SFSHH29 pKa = 5.92ATYY32 pKa = 7.61TTLPLPTDD40 pKa = 3.7IYY42 pKa = 11.49QMLRR46 pKa = 11.84CVEE49 pKa = 4.28VYY51 pKa = 10.66LEE53 pKa = 4.01RR54 pKa = 11.84LYY56 pKa = 10.69TADD59 pKa = 2.84VTRR62 pKa = 11.84YY63 pKa = 9.19VDD65 pKa = 3.37VVSNYY70 pKa = 9.92RR71 pKa = 11.84KK72 pKa = 8.01TSVARR77 pKa = 11.84AVPHH81 pKa = 5.96IFVVNSMEE89 pKa = 4.2KK90 pKa = 10.58FSTSYY95 pKa = 9.62STLLSKK101 pKa = 10.82SPHH104 pKa = 7.16DD105 pKa = 4.0YY106 pKa = 10.54VSSDD110 pKa = 3.22VTVVSNPTSQDD121 pKa = 2.71IIRR124 pKa = 11.84SIRR127 pKa = 11.84AHH129 pKa = 5.65KK130 pKa = 10.22HH131 pKa = 3.51VVCNVVPIGDD141 pKa = 3.77FSDD144 pKa = 5.02AILMPNASISDD155 pKa = 3.7CVKK158 pKa = 8.48YY159 pKa = 10.76LRR161 pKa = 11.84RR162 pKa = 11.84NFHH165 pKa = 6.14ALLRR169 pKa = 11.84RR170 pKa = 11.84LSFGHH175 pKa = 6.37GCYY178 pKa = 10.1SCSNVLQLSLPPWLSVLASGWQRR201 pKa = 11.84FKK203 pKa = 10.66PTGSDD208 pKa = 2.63TDD210 pKa = 3.95FAQFYY215 pKa = 7.28LTPARR220 pKa = 11.84PQLSVINSISDD231 pKa = 3.42LSDD234 pKa = 4.07FIINRR239 pKa = 11.84LQANVDD245 pKa = 3.3VVYY248 pKa = 10.69FDD250 pKa = 6.15DD251 pKa = 4.97VLTTDD256 pKa = 3.43GRR258 pKa = 11.84TIAEE262 pKa = 4.63KK263 pKa = 10.39IRR265 pKa = 11.84ATGAQFNVARR275 pKa = 11.84DD276 pKa = 3.61MTHH279 pKa = 6.76FNIIKK284 pKa = 9.62QRR286 pKa = 11.84YY287 pKa = 8.28LEE289 pKa = 3.94QTSIRR294 pKa = 11.84RR295 pKa = 11.84RR296 pKa = 11.84RR297 pKa = 11.84QHH299 pKa = 6.42KK300 pKa = 10.37KK301 pKa = 9.19EE302 pKa = 3.71LHH304 pKa = 5.72ASDD307 pKa = 3.95RR308 pKa = 11.84KK309 pKa = 9.36QNIVARR315 pKa = 11.84VFPPVYY321 pKa = 10.33LVDD324 pKa = 4.47LSVSIRR330 pKa = 11.84AWNSAMPTQQQRR342 pKa = 11.84RR343 pKa = 11.84LSRR346 pKa = 11.84VYY348 pKa = 10.87SDD350 pKa = 4.68LVNLSKK356 pKa = 10.93RR357 pKa = 11.84PLTVLHH363 pKa = 6.44MIAWNGPSVVSTDD376 pKa = 2.91SDD378 pKa = 3.43VWQDD382 pKa = 3.23FTEE385 pKa = 3.98WLFCVVQNVDD395 pKa = 2.79EE396 pKa = 4.5SMARR400 pKa = 11.84LLTAIPYY407 pKa = 8.59MSVEE411 pKa = 4.13IKK413 pKa = 10.49ISGSGFGTWIDD424 pKa = 3.5LRR426 pKa = 11.84NWTEE430 pKa = 4.05GFSLSKK436 pKa = 10.41ILVGRR441 pKa = 11.84TYY443 pKa = 10.47MVGKK447 pKa = 9.93KK448 pKa = 10.13NSGKK452 pKa = 10.94GMIGKK457 pKa = 8.64LIWRR461 pKa = 11.84LGVPVVDD468 pKa = 3.5SDD470 pKa = 4.06DD471 pKa = 3.7YY472 pKa = 11.99GRR474 pKa = 11.84VLLIAEE480 pKa = 4.21NTGVSLEE487 pKa = 4.19DD488 pKa = 3.29AVRR491 pKa = 11.84CHH493 pKa = 6.59FSRR496 pKa = 11.84SYY498 pKa = 11.52ADD500 pKa = 4.03RR501 pKa = 11.84DD502 pKa = 3.7CAPTLFEE509 pKa = 5.11TIMDD513 pKa = 5.3DD514 pKa = 2.82IVTRR518 pKa = 11.84LSIKK522 pKa = 9.96RR523 pKa = 11.84YY524 pKa = 9.39SIPEE528 pKa = 3.78VDD530 pKa = 4.58HH531 pKa = 6.89PALMMFGHH539 pKa = 7.57AYY541 pKa = 10.45DD542 pKa = 4.53EE543 pKa = 4.18LTKK546 pKa = 10.25KK547 pKa = 10.55YY548 pKa = 10.37KK549 pKa = 10.37YY550 pKa = 10.74ADD552 pKa = 3.64FEE554 pKa = 5.14RR555 pKa = 11.84MVRR558 pKa = 11.84SFISHH563 pKa = 6.5HH564 pKa = 5.83GVRR567 pKa = 11.84APDD570 pKa = 3.68GSLLKK575 pKa = 10.59VDD577 pKa = 4.08DD578 pKa = 4.2RR579 pKa = 11.84FVFSTHH585 pKa = 6.2CSEE588 pKa = 5.02EE589 pKa = 3.95ATQVLGANYY598 pKa = 8.8MFQLSTAIDD607 pKa = 3.85SYY609 pKa = 11.72VGVLLRR615 pKa = 11.84GQHH618 pKa = 6.52DD619 pKa = 3.83NSISEE624 pKa = 4.46LMLAVYY630 pKa = 10.0YY631 pKa = 10.69DD632 pKa = 4.57RR633 pKa = 11.84IHH635 pKa = 7.83VNIFDD640 pKa = 4.57LVPTGVVCTVLRR652 pKa = 11.84TGLARR657 pKa = 11.84LVPKK661 pKa = 10.64AA662 pKa = 3.7
Molecular weight: 74.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7719 |
269 |
1360 |
643.3 |
71.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.931 ± 0.282 | 1.555 ± 0.132 |
6.18 ± 0.334 | 4.508 ± 0.245 |
4.431 ± 0.228 | 6.141 ± 0.356 |
2.902 ± 0.215 | 5.247 ± 0.144 |
2.734 ± 0.294 | 8.835 ± 0.287 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.902 ± 0.207 | 3.822 ± 0.16 |
5.299 ± 0.215 | 3.187 ± 0.173 |
6.996 ± 0.278 | 8.317 ± 0.579 |
6.27 ± 0.256 | 8.796 ± 0.505 |
1.296 ± 0.131 | 3.64 ± 0.235 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
