
Streptomyces sp. TSRI0107
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; unclassified Streptomyces
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
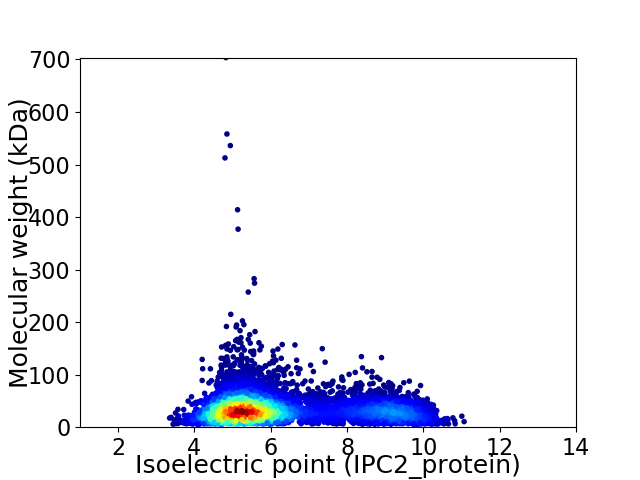
Virtual 2D-PAGE plot for 7155 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Q5L7F5|A0A1Q5L7F5_9ACTN ABC transporter substrate-binding protein OS=Streptomyces sp. TSRI0107 OX=1703942 GN=AMK31_00400 PE=4 SV=1
MM1 pKa = 7.81TDD3 pKa = 3.42DD4 pKa = 3.32QHH6 pKa = 9.61GEE8 pKa = 4.06GTPQGGGRR16 pKa = 11.84WDD18 pKa = 4.22PLPQGDD24 pKa = 4.33YY25 pKa = 11.28DD26 pKa = 4.37DD27 pKa = 4.9GATAFVQLPEE37 pKa = 4.38GGIDD41 pKa = 3.5ALLAASEE48 pKa = 4.73SPLAAPGHH56 pKa = 6.43GYY58 pKa = 9.86VPPPITVTPATTAGSDD74 pKa = 3.59PAATGAWSMPAEE86 pKa = 4.68GAQWPDD92 pKa = 3.61PEE94 pKa = 5.62AVPQQDD100 pKa = 3.24AAHH103 pKa = 6.95DD104 pKa = 3.9RR105 pKa = 11.84FTYY108 pKa = 10.57DD109 pKa = 4.04PGATGQWTYY118 pKa = 11.82AEE120 pKa = 4.31PPEE123 pKa = 4.46TYY125 pKa = 10.18EE126 pKa = 4.24EE127 pKa = 4.14QPEE130 pKa = 4.03ALPAPGHH137 pKa = 6.65DD138 pKa = 3.68VTGQWSIPVAGGDD151 pKa = 3.84LPDD154 pKa = 3.81EE155 pKa = 4.7SGEE158 pKa = 4.22FTTSALVEE166 pKa = 4.23QWGGTPPATLPGGAPAPWATEE187 pKa = 3.99GAGQAWAQQATAGDD201 pKa = 4.43GGRR204 pKa = 11.84HH205 pKa = 5.52PQDD208 pKa = 3.58HH209 pKa = 6.5PQSHH213 pKa = 6.32SEE215 pKa = 3.75AHH217 pKa = 6.49AEE219 pKa = 3.82PTPAHH224 pKa = 5.76GMYY227 pKa = 10.21VGHH230 pKa = 6.97LPEE233 pKa = 6.36AEE235 pKa = 4.37AEE237 pKa = 4.25AEE239 pKa = 4.31SAEE242 pKa = 4.2APAPEE247 pKa = 5.21AGEE250 pKa = 4.12AGEE253 pKa = 4.95AGGAPEE259 pKa = 4.5TPGASDD265 pKa = 3.14TAGAGEE271 pKa = 4.07AVEE274 pKa = 4.31AVSGAVEE281 pKa = 4.35TPEE284 pKa = 4.04GTEE287 pKa = 4.01PTGMAPDD294 pKa = 4.09APTGGEE300 pKa = 4.13SAAEE304 pKa = 4.38GPAEE308 pKa = 4.33AEE310 pKa = 4.23AEE312 pKa = 4.32VLPDD316 pKa = 4.41AEE318 pKa = 4.47PAQAAGTPPPPSDD331 pKa = 3.25AAPEE335 pKa = 4.05QEE337 pKa = 4.84HH338 pKa = 6.56APEE341 pKa = 4.37PAAAPDD347 pKa = 3.67AAEE350 pKa = 5.33DD351 pKa = 4.11PPALPGEE358 pKa = 4.17EE359 pKa = 4.74HH360 pKa = 6.93PLASYY365 pKa = 9.32VLRR368 pKa = 11.84VNGGDD373 pKa = 4.29RR374 pKa = 11.84PVTDD378 pKa = 3.0AWIGEE383 pKa = 4.15SLLYY387 pKa = 10.18VLRR390 pKa = 11.84EE391 pKa = 3.94RR392 pKa = 11.84LGLAGAKK399 pKa = 9.72DD400 pKa = 3.75GCSQGEE406 pKa = 4.12CGACNVQVDD415 pKa = 3.94GRR417 pKa = 11.84LVASCLVPAVTAAGSEE433 pKa = 4.05VRR435 pKa = 11.84TVEE438 pKa = 3.92GLAVDD443 pKa = 4.62GQPSDD448 pKa = 3.61VQRR451 pKa = 11.84ALARR455 pKa = 11.84CGAVQCGFCVPGMAMTVHH473 pKa = 7.27DD474 pKa = 5.16LLEE477 pKa = 4.78GNPAPTEE484 pKa = 4.08LEE486 pKa = 4.08TRR488 pKa = 11.84QALCGNLCRR497 pKa = 11.84CSGYY501 pKa = 10.16RR502 pKa = 11.84AVVDD506 pKa = 3.87AVQEE510 pKa = 4.4VVAEE514 pKa = 4.29RR515 pKa = 11.84DD516 pKa = 2.96AHH518 pKa = 5.75AASDD522 pKa = 3.55AAEE525 pKa = 4.13TDD527 pKa = 3.71EE528 pKa = 5.81ARR530 pKa = 11.84IPHH533 pKa = 5.5QAGPGAGGVNPSAFEE548 pKa = 3.81AFQPHH553 pKa = 7.1DD554 pKa = 3.79PSFGQGPGDD563 pKa = 3.72GQGQGQGQDD572 pKa = 3.28GGQVV576 pKa = 2.88
MM1 pKa = 7.81TDD3 pKa = 3.42DD4 pKa = 3.32QHH6 pKa = 9.61GEE8 pKa = 4.06GTPQGGGRR16 pKa = 11.84WDD18 pKa = 4.22PLPQGDD24 pKa = 4.33YY25 pKa = 11.28DD26 pKa = 4.37DD27 pKa = 4.9GATAFVQLPEE37 pKa = 4.38GGIDD41 pKa = 3.5ALLAASEE48 pKa = 4.73SPLAAPGHH56 pKa = 6.43GYY58 pKa = 9.86VPPPITVTPATTAGSDD74 pKa = 3.59PAATGAWSMPAEE86 pKa = 4.68GAQWPDD92 pKa = 3.61PEE94 pKa = 5.62AVPQQDD100 pKa = 3.24AAHH103 pKa = 6.95DD104 pKa = 3.9RR105 pKa = 11.84FTYY108 pKa = 10.57DD109 pKa = 4.04PGATGQWTYY118 pKa = 11.82AEE120 pKa = 4.31PPEE123 pKa = 4.46TYY125 pKa = 10.18EE126 pKa = 4.24EE127 pKa = 4.14QPEE130 pKa = 4.03ALPAPGHH137 pKa = 6.65DD138 pKa = 3.68VTGQWSIPVAGGDD151 pKa = 3.84LPDD154 pKa = 3.81EE155 pKa = 4.7SGEE158 pKa = 4.22FTTSALVEE166 pKa = 4.23QWGGTPPATLPGGAPAPWATEE187 pKa = 3.99GAGQAWAQQATAGDD201 pKa = 4.43GGRR204 pKa = 11.84HH205 pKa = 5.52PQDD208 pKa = 3.58HH209 pKa = 6.5PQSHH213 pKa = 6.32SEE215 pKa = 3.75AHH217 pKa = 6.49AEE219 pKa = 3.82PTPAHH224 pKa = 5.76GMYY227 pKa = 10.21VGHH230 pKa = 6.97LPEE233 pKa = 6.36AEE235 pKa = 4.37AEE237 pKa = 4.25AEE239 pKa = 4.31SAEE242 pKa = 4.2APAPEE247 pKa = 5.21AGEE250 pKa = 4.12AGEE253 pKa = 4.95AGGAPEE259 pKa = 4.5TPGASDD265 pKa = 3.14TAGAGEE271 pKa = 4.07AVEE274 pKa = 4.31AVSGAVEE281 pKa = 4.35TPEE284 pKa = 4.04GTEE287 pKa = 4.01PTGMAPDD294 pKa = 4.09APTGGEE300 pKa = 4.13SAAEE304 pKa = 4.38GPAEE308 pKa = 4.33AEE310 pKa = 4.23AEE312 pKa = 4.32VLPDD316 pKa = 4.41AEE318 pKa = 4.47PAQAAGTPPPPSDD331 pKa = 3.25AAPEE335 pKa = 4.05QEE337 pKa = 4.84HH338 pKa = 6.56APEE341 pKa = 4.37PAAAPDD347 pKa = 3.67AAEE350 pKa = 5.33DD351 pKa = 4.11PPALPGEE358 pKa = 4.17EE359 pKa = 4.74HH360 pKa = 6.93PLASYY365 pKa = 9.32VLRR368 pKa = 11.84VNGGDD373 pKa = 4.29RR374 pKa = 11.84PVTDD378 pKa = 3.0AWIGEE383 pKa = 4.15SLLYY387 pKa = 10.18VLRR390 pKa = 11.84EE391 pKa = 3.94RR392 pKa = 11.84LGLAGAKK399 pKa = 9.72DD400 pKa = 3.75GCSQGEE406 pKa = 4.12CGACNVQVDD415 pKa = 3.94GRR417 pKa = 11.84LVASCLVPAVTAAGSEE433 pKa = 4.05VRR435 pKa = 11.84TVEE438 pKa = 3.92GLAVDD443 pKa = 4.62GQPSDD448 pKa = 3.61VQRR451 pKa = 11.84ALARR455 pKa = 11.84CGAVQCGFCVPGMAMTVHH473 pKa = 7.27DD474 pKa = 5.16LLEE477 pKa = 4.78GNPAPTEE484 pKa = 4.08LEE486 pKa = 4.08TRR488 pKa = 11.84QALCGNLCRR497 pKa = 11.84CSGYY501 pKa = 10.16RR502 pKa = 11.84AVVDD506 pKa = 3.87AVQEE510 pKa = 4.4VVAEE514 pKa = 4.29RR515 pKa = 11.84DD516 pKa = 2.96AHH518 pKa = 5.75AASDD522 pKa = 3.55AAEE525 pKa = 4.13TDD527 pKa = 3.71EE528 pKa = 5.81ARR530 pKa = 11.84IPHH533 pKa = 5.5QAGPGAGGVNPSAFEE548 pKa = 3.81AFQPHH553 pKa = 7.1DD554 pKa = 3.79PSFGQGPGDD563 pKa = 3.72GQGQGQGQDD572 pKa = 3.28GGQVV576 pKa = 2.88
Molecular weight: 57.88 kDa
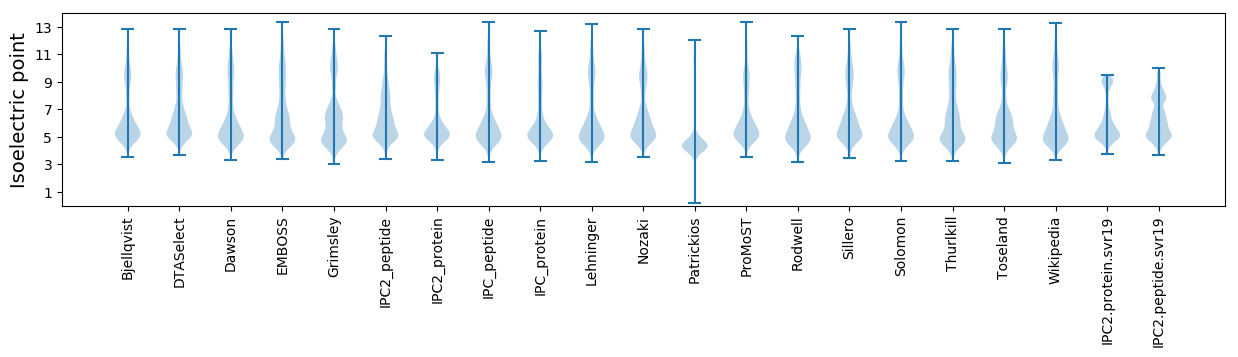
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Q5L143|A0A1Q5L143_9ACTN zf-HC2 domain-containing protein OS=Streptomyces sp. TSRI0107 OX=1703942 GN=AMK31_10425 PE=4 SV=1
MM1 pKa = 7.17AAVRR5 pKa = 11.84VVRR8 pKa = 11.84VVPAAPVGRR17 pKa = 11.84VAAASPAVPAAAVAPVVRR35 pKa = 11.84AVAAASPVVRR45 pKa = 11.84VLPAVAAASAAAVAVPASAAVPAVPVAVVARR76 pKa = 11.84RR77 pKa = 11.84ARR79 pKa = 11.84SVARR83 pKa = 11.84VVPRR87 pKa = 11.84VAAASRR93 pKa = 11.84SGRR96 pKa = 11.84GARR99 pKa = 11.84STRR102 pKa = 11.84PCRR105 pKa = 11.84PRR107 pKa = 11.84ASAAA111 pKa = 3.29
MM1 pKa = 7.17AAVRR5 pKa = 11.84VVRR8 pKa = 11.84VVPAAPVGRR17 pKa = 11.84VAAASPAVPAAAVAPVVRR35 pKa = 11.84AVAAASPVVRR45 pKa = 11.84VLPAVAAASAAAVAVPASAAVPAVPVAVVARR76 pKa = 11.84RR77 pKa = 11.84ARR79 pKa = 11.84SVARR83 pKa = 11.84VVPRR87 pKa = 11.84VAAASRR93 pKa = 11.84SGRR96 pKa = 11.84GARR99 pKa = 11.84STRR102 pKa = 11.84PCRR105 pKa = 11.84PRR107 pKa = 11.84ASAAA111 pKa = 3.29
Molecular weight: 10.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2402202 |
37 |
6726 |
335.7 |
35.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.696 ± 0.051 | 0.769 ± 0.006 |
6.124 ± 0.023 | 5.864 ± 0.029 |
2.709 ± 0.015 | 9.467 ± 0.032 |
2.309 ± 0.014 | 3.01 ± 0.02 |
2.067 ± 0.022 | 10.441 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.681 ± 0.012 | 1.698 ± 0.017 |
6.165 ± 0.026 | 2.679 ± 0.017 |
8.19 ± 0.035 | 4.79 ± 0.025 |
6.138 ± 0.025 | 8.549 ± 0.025 |
1.545 ± 0.012 | 2.111 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
