
Alistipes sp. CHKCI003
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; unclassified Alistipes
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
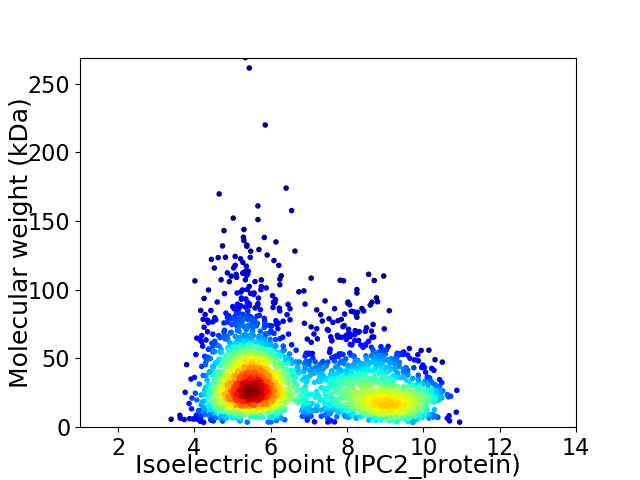
Virtual 2D-PAGE plot for 2504 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A143XFJ7|A0A143XFJ7_9BACT Outer membrane lipoprotein Blc OS=Alistipes sp. CHKCI003 OX=1780376 GN=blc PE=3 SV=1
MM1 pKa = 7.44SIKK4 pKa = 10.55HH5 pKa = 6.32LFYY8 pKa = 11.19ALAVSGMLSGAVSCADD24 pKa = 4.2DD25 pKa = 3.86EE26 pKa = 5.04PEE28 pKa = 4.25TEE30 pKa = 4.3VVPVDD35 pKa = 3.67FTLRR39 pKa = 11.84LNMPISLTDD48 pKa = 3.7PKK50 pKa = 10.13LTSVDD55 pKa = 3.73CAFTRR60 pKa = 11.84VEE62 pKa = 4.04TGDD65 pKa = 3.35EE66 pKa = 3.95VRR68 pKa = 11.84ITEE71 pKa = 4.11FTEE74 pKa = 4.54AEE76 pKa = 3.93TGAYY80 pKa = 8.86EE81 pKa = 4.34VVATLEE87 pKa = 4.0EE88 pKa = 4.29GTYY91 pKa = 10.53NIAVSGQIAYY101 pKa = 8.64TLDD104 pKa = 3.41GNTAEE109 pKa = 4.78RR110 pKa = 11.84DD111 pKa = 3.3IEE113 pKa = 4.01ASRR116 pKa = 11.84EE117 pKa = 4.09GVAVSAASDD126 pKa = 3.5VLTIEE131 pKa = 4.29TTVAAMKK138 pKa = 10.57SDD140 pKa = 3.48DD141 pKa = 3.92FVIEE145 pKa = 4.48EE146 pKa = 3.91IFYY149 pKa = 10.05TGTEE153 pKa = 4.28TPEE156 pKa = 3.66GMPYY160 pKa = 10.69YY161 pKa = 10.36GDD163 pKa = 3.14QYY165 pKa = 11.5MKK167 pKa = 9.5ITNNSDD173 pKa = 2.88RR174 pKa = 11.84TLYY177 pKa = 11.13ADD179 pKa = 3.48GLVIFKK185 pKa = 10.47SAFMTVEE192 pKa = 4.14KK193 pKa = 10.8YY194 pKa = 10.79DD195 pKa = 3.73YY196 pKa = 10.28TPDD199 pKa = 3.07IMSDD203 pKa = 3.73YY204 pKa = 10.72FSVDD208 pKa = 2.47SGLILPGSGTDD219 pKa = 3.54YY220 pKa = 10.77PVAPGASVVIADD232 pKa = 3.73TAIDD236 pKa = 3.5HH237 pKa = 6.64TEE239 pKa = 3.98FNSLSLDD246 pKa = 3.42LSGATFEE253 pKa = 4.74YY254 pKa = 10.76FNTLDD259 pKa = 5.37DD260 pKa = 4.6EE261 pKa = 6.0DD262 pKa = 4.46MDD264 pKa = 4.78NPDD267 pKa = 3.64VPNVTTLFADD277 pKa = 5.79DD278 pKa = 3.76FWTWHH283 pKa = 5.07NRR285 pKa = 11.84GFYY288 pKa = 9.51TYY290 pKa = 10.71AIGRR294 pKa = 11.84LGEE297 pKa = 4.24GVTAEE302 pKa = 4.57SFASDD307 pKa = 3.38YY308 pKa = 10.95AYY310 pKa = 9.23TVEE313 pKa = 4.21YY314 pKa = 10.75EE315 pKa = 4.39MVVPDD320 pKa = 4.02YY321 pKa = 10.53GTFPMSDD328 pKa = 4.6DD329 pKa = 3.3YY330 pKa = 11.61WKK332 pKa = 11.1VPNSWIIDD340 pKa = 3.63AVNLSIEE347 pKa = 4.4EE348 pKa = 4.06SFAWIVTSPLLDD360 pKa = 4.7AGWTYY365 pKa = 10.84CGKK368 pKa = 10.04IDD370 pKa = 4.48GDD372 pKa = 3.52EE373 pKa = 4.11TRR375 pKa = 11.84FGKK378 pKa = 8.99SVRR381 pKa = 11.84RR382 pKa = 11.84KK383 pKa = 9.58SVSTTSSPYY392 pKa = 11.28YY393 pKa = 9.79MDD395 pKa = 4.75TNDD398 pKa = 3.29STADD402 pKa = 4.54FIPEE406 pKa = 4.32TSPSLKK412 pKa = 10.61NN413 pKa = 3.33
MM1 pKa = 7.44SIKK4 pKa = 10.55HH5 pKa = 6.32LFYY8 pKa = 11.19ALAVSGMLSGAVSCADD24 pKa = 4.2DD25 pKa = 3.86EE26 pKa = 5.04PEE28 pKa = 4.25TEE30 pKa = 4.3VVPVDD35 pKa = 3.67FTLRR39 pKa = 11.84LNMPISLTDD48 pKa = 3.7PKK50 pKa = 10.13LTSVDD55 pKa = 3.73CAFTRR60 pKa = 11.84VEE62 pKa = 4.04TGDD65 pKa = 3.35EE66 pKa = 3.95VRR68 pKa = 11.84ITEE71 pKa = 4.11FTEE74 pKa = 4.54AEE76 pKa = 3.93TGAYY80 pKa = 8.86EE81 pKa = 4.34VVATLEE87 pKa = 4.0EE88 pKa = 4.29GTYY91 pKa = 10.53NIAVSGQIAYY101 pKa = 8.64TLDD104 pKa = 3.41GNTAEE109 pKa = 4.78RR110 pKa = 11.84DD111 pKa = 3.3IEE113 pKa = 4.01ASRR116 pKa = 11.84EE117 pKa = 4.09GVAVSAASDD126 pKa = 3.5VLTIEE131 pKa = 4.29TTVAAMKK138 pKa = 10.57SDD140 pKa = 3.48DD141 pKa = 3.92FVIEE145 pKa = 4.48EE146 pKa = 3.91IFYY149 pKa = 10.05TGTEE153 pKa = 4.28TPEE156 pKa = 3.66GMPYY160 pKa = 10.69YY161 pKa = 10.36GDD163 pKa = 3.14QYY165 pKa = 11.5MKK167 pKa = 9.5ITNNSDD173 pKa = 2.88RR174 pKa = 11.84TLYY177 pKa = 11.13ADD179 pKa = 3.48GLVIFKK185 pKa = 10.47SAFMTVEE192 pKa = 4.14KK193 pKa = 10.8YY194 pKa = 10.79DD195 pKa = 3.73YY196 pKa = 10.28TPDD199 pKa = 3.07IMSDD203 pKa = 3.73YY204 pKa = 10.72FSVDD208 pKa = 2.47SGLILPGSGTDD219 pKa = 3.54YY220 pKa = 10.77PVAPGASVVIADD232 pKa = 3.73TAIDD236 pKa = 3.5HH237 pKa = 6.64TEE239 pKa = 3.98FNSLSLDD246 pKa = 3.42LSGATFEE253 pKa = 4.74YY254 pKa = 10.76FNTLDD259 pKa = 5.37DD260 pKa = 4.6EE261 pKa = 6.0DD262 pKa = 4.46MDD264 pKa = 4.78NPDD267 pKa = 3.64VPNVTTLFADD277 pKa = 5.79DD278 pKa = 3.76FWTWHH283 pKa = 5.07NRR285 pKa = 11.84GFYY288 pKa = 9.51TYY290 pKa = 10.71AIGRR294 pKa = 11.84LGEE297 pKa = 4.24GVTAEE302 pKa = 4.57SFASDD307 pKa = 3.38YY308 pKa = 10.95AYY310 pKa = 9.23TVEE313 pKa = 4.21YY314 pKa = 10.75EE315 pKa = 4.39MVVPDD320 pKa = 4.02YY321 pKa = 10.53GTFPMSDD328 pKa = 4.6DD329 pKa = 3.3YY330 pKa = 11.61WKK332 pKa = 11.1VPNSWIIDD340 pKa = 3.63AVNLSIEE347 pKa = 4.4EE348 pKa = 4.06SFAWIVTSPLLDD360 pKa = 4.7AGWTYY365 pKa = 10.84CGKK368 pKa = 10.04IDD370 pKa = 4.48GDD372 pKa = 3.52EE373 pKa = 4.11TRR375 pKa = 11.84FGKK378 pKa = 8.99SVRR381 pKa = 11.84RR382 pKa = 11.84KK383 pKa = 9.58SVSTTSSPYY392 pKa = 11.28YY393 pKa = 9.79MDD395 pKa = 4.75TNDD398 pKa = 3.29STADD402 pKa = 4.54FIPEE406 pKa = 4.32TSPSLKK412 pKa = 10.61NN413 pKa = 3.33
Molecular weight: 45.51 kDa
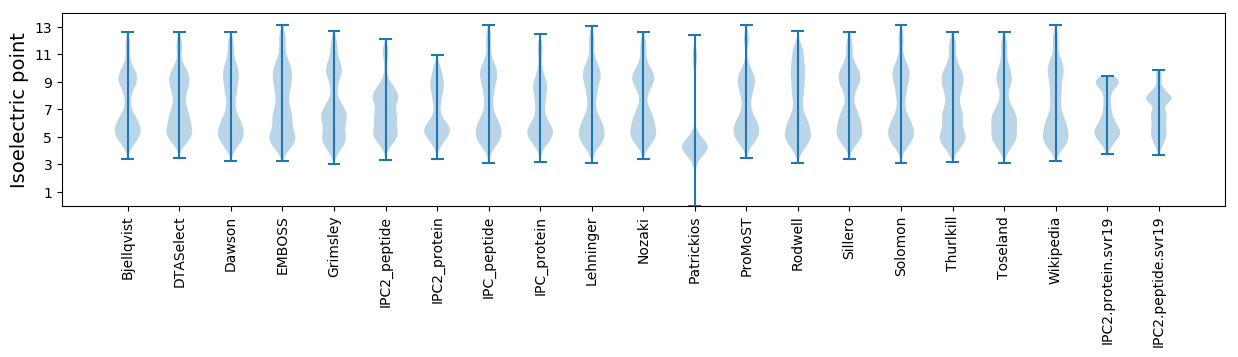
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A143XTZ2|A0A143XTZ2_9BACT Phosphoethanolamine transferase EptB OS=Alistipes sp. CHKCI003 OX=1780376 GN=eptB PE=4 SV=1
MM1 pKa = 7.84PNGKK5 pKa = 8.55KK6 pKa = 9.72HH7 pKa = 6.1KK8 pKa = 7.52RR9 pKa = 11.84HH10 pKa = 5.93KK11 pKa = 9.95MATHH15 pKa = 6.02KK16 pKa = 10.34RR17 pKa = 11.84KK18 pKa = 9.84KK19 pKa = 9.28RR20 pKa = 11.84LRR22 pKa = 11.84KK23 pKa = 9.26NRR25 pKa = 11.84HH26 pKa = 4.71KK27 pKa = 10.87KK28 pKa = 9.36KK29 pKa = 10.77
MM1 pKa = 7.84PNGKK5 pKa = 8.55KK6 pKa = 9.72HH7 pKa = 6.1KK8 pKa = 7.52RR9 pKa = 11.84HH10 pKa = 5.93KK11 pKa = 9.95MATHH15 pKa = 6.02KK16 pKa = 10.34RR17 pKa = 11.84KK18 pKa = 9.84KK19 pKa = 9.28RR20 pKa = 11.84LRR22 pKa = 11.84KK23 pKa = 9.26NRR25 pKa = 11.84HH26 pKa = 4.71KK27 pKa = 10.87KK28 pKa = 9.36KK29 pKa = 10.77
Molecular weight: 3.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
798870 |
29 |
2392 |
319.0 |
35.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.81 ± 0.071 | 1.41 ± 0.021 |
5.645 ± 0.04 | 6.641 ± 0.049 |
4.209 ± 0.032 | 7.607 ± 0.048 |
1.79 ± 0.021 | 5.601 ± 0.042 |
4.53 ± 0.047 | 8.952 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.476 ± 0.024 | 3.485 ± 0.036 |
4.418 ± 0.033 | 2.824 ± 0.031 |
7.569 ± 0.079 | 5.723 ± 0.039 |
5.577 ± 0.045 | 6.865 ± 0.044 |
1.106 ± 0.017 | 3.76 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
