
Heliophilum fasciatum
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Heliobacteriaceae; Heliophilum
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
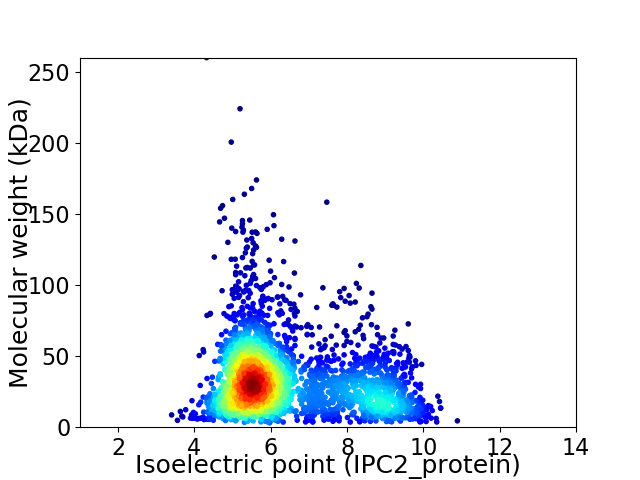
Virtual 2D-PAGE plot for 2911 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
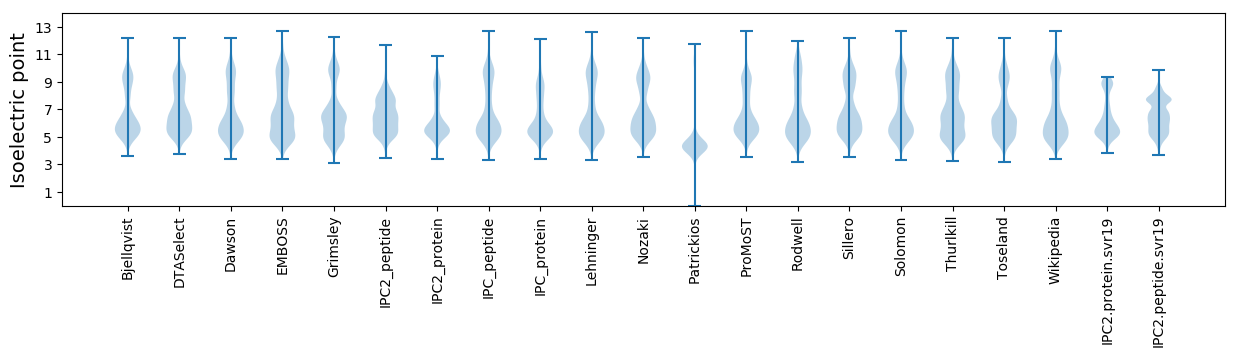
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R2R7R9|A0A4R2R7R9_9FIRM Type III restriction/modification enzyme methylation subunit OS=Heliophilum fasciatum OX=35700 GN=EDD73_1572 PE=4 SV=1
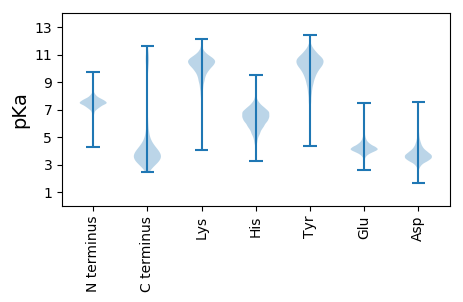
MM1 pKa = 7.75LDD3 pKa = 3.77DD4 pKa = 4.96RR5 pKa = 11.84DD6 pKa = 3.73YY7 pKa = 11.69SADD10 pKa = 3.55GFHH13 pKa = 6.83ATDD16 pKa = 5.19ADD18 pKa = 3.85FTDD21 pKa = 3.91NYY23 pKa = 10.38IDD25 pKa = 5.32AEE27 pKa = 4.09PSYY30 pKa = 10.75QDD32 pKa = 2.87EE33 pKa = 4.52GFYY36 pKa = 11.11DD37 pKa = 5.2GIPRR41 pKa = 11.84II42 pKa = 4.07
MM1 pKa = 7.75LDD3 pKa = 3.77DD4 pKa = 4.96RR5 pKa = 11.84DD6 pKa = 3.73YY7 pKa = 11.69SADD10 pKa = 3.55GFHH13 pKa = 6.83ATDD16 pKa = 5.19ADD18 pKa = 3.85FTDD21 pKa = 3.91NYY23 pKa = 10.38IDD25 pKa = 5.32AEE27 pKa = 4.09PSYY30 pKa = 10.75QDD32 pKa = 2.87EE33 pKa = 4.52GFYY36 pKa = 11.11DD37 pKa = 5.2GIPRR41 pKa = 11.84II42 pKa = 4.07
Molecular weight: 4.82 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R2RY77|A0A4R2RY77_9FIRM RND family efflux transporter MFP subunit OS=Heliophilum fasciatum OX=35700 GN=EDD73_11226 PE=3 SV=1
MM1 pKa = 7.89PYY3 pKa = 9.71IIHH6 pKa = 6.58RR7 pKa = 11.84NRR9 pKa = 11.84RR10 pKa = 11.84IFFEE14 pKa = 4.07EE15 pKa = 4.32RR16 pKa = 11.84GDD18 pKa = 3.9GAPLLLIHH26 pKa = 6.52GVAVSHH32 pKa = 6.65KK33 pKa = 8.4MWAPQLAVLARR44 pKa = 11.84SYY46 pKa = 11.1RR47 pKa = 11.84VIAPDD52 pKa = 3.3LRR54 pKa = 11.84GAGKK58 pKa = 10.47SSGLSWQSRR67 pKa = 11.84VADD70 pKa = 4.35LVADD74 pKa = 5.31LIALLDD80 pKa = 3.75RR81 pKa = 11.84LAIEE85 pKa = 4.13QVAVCGLSFGGVVAQEE101 pKa = 3.74LVLRR105 pKa = 11.84YY106 pKa = 8.96PEE108 pKa = 4.22RR109 pKa = 11.84CAMLVLVDD117 pKa = 4.07TFSEE121 pKa = 4.14LRR123 pKa = 11.84PGISQEE129 pKa = 3.93GALWLATWLGVPLFLMPKK147 pKa = 8.07RR148 pKa = 11.84WMLARR153 pKa = 11.84MKK155 pKa = 10.12EE156 pKa = 4.56SYY158 pKa = 10.65GKK160 pKa = 9.48WPVACRR166 pKa = 11.84CLTAEE171 pKa = 3.93IPKK174 pKa = 9.69IRR176 pKa = 11.84GMEE179 pKa = 4.05AVKK182 pKa = 9.67MRR184 pKa = 11.84WMMRR188 pKa = 11.84GLRR191 pKa = 11.84LTPRR195 pKa = 11.84LAAIHH200 pKa = 6.28CPCLGIVGGNTALGIALMRR219 pKa = 11.84RR220 pKa = 11.84VIEE223 pKa = 4.92AIPGAKK229 pKa = 9.69LRR231 pKa = 11.84VVPHH235 pKa = 6.68AFDD238 pKa = 4.03PTSLCQPAIFNRR250 pKa = 11.84LVDD253 pKa = 3.9EE254 pKa = 6.02FLAPWQEE261 pKa = 3.36RR262 pKa = 11.84WAARR266 pKa = 11.84SSEE269 pKa = 3.91RR270 pKa = 11.84FLRR273 pKa = 11.84KK274 pKa = 9.28ISARR278 pKa = 11.84ARR280 pKa = 11.84NRR282 pKa = 11.84QSICRR287 pKa = 11.84SRR289 pKa = 11.84WEE291 pKa = 3.96PSFITLGVGQRR302 pKa = 11.84NAKK305 pKa = 10.36DD306 pKa = 3.23GAEE309 pKa = 3.94RR310 pKa = 3.8
MM1 pKa = 7.89PYY3 pKa = 9.71IIHH6 pKa = 6.58RR7 pKa = 11.84NRR9 pKa = 11.84RR10 pKa = 11.84IFFEE14 pKa = 4.07EE15 pKa = 4.32RR16 pKa = 11.84GDD18 pKa = 3.9GAPLLLIHH26 pKa = 6.52GVAVSHH32 pKa = 6.65KK33 pKa = 8.4MWAPQLAVLARR44 pKa = 11.84SYY46 pKa = 11.1RR47 pKa = 11.84VIAPDD52 pKa = 3.3LRR54 pKa = 11.84GAGKK58 pKa = 10.47SSGLSWQSRR67 pKa = 11.84VADD70 pKa = 4.35LVADD74 pKa = 5.31LIALLDD80 pKa = 3.75RR81 pKa = 11.84LAIEE85 pKa = 4.13QVAVCGLSFGGVVAQEE101 pKa = 3.74LVLRR105 pKa = 11.84YY106 pKa = 8.96PEE108 pKa = 4.22RR109 pKa = 11.84CAMLVLVDD117 pKa = 4.07TFSEE121 pKa = 4.14LRR123 pKa = 11.84PGISQEE129 pKa = 3.93GALWLATWLGVPLFLMPKK147 pKa = 8.07RR148 pKa = 11.84WMLARR153 pKa = 11.84MKK155 pKa = 10.12EE156 pKa = 4.56SYY158 pKa = 10.65GKK160 pKa = 9.48WPVACRR166 pKa = 11.84CLTAEE171 pKa = 3.93IPKK174 pKa = 9.69IRR176 pKa = 11.84GMEE179 pKa = 4.05AVKK182 pKa = 9.67MRR184 pKa = 11.84WMMRR188 pKa = 11.84GLRR191 pKa = 11.84LTPRR195 pKa = 11.84LAAIHH200 pKa = 6.28CPCLGIVGGNTALGIALMRR219 pKa = 11.84RR220 pKa = 11.84VIEE223 pKa = 4.92AIPGAKK229 pKa = 9.69LRR231 pKa = 11.84VVPHH235 pKa = 6.68AFDD238 pKa = 4.03PTSLCQPAIFNRR250 pKa = 11.84LVDD253 pKa = 3.9EE254 pKa = 6.02FLAPWQEE261 pKa = 3.36RR262 pKa = 11.84WAARR266 pKa = 11.84SSEE269 pKa = 3.91RR270 pKa = 11.84FLRR273 pKa = 11.84KK274 pKa = 9.28ISARR278 pKa = 11.84ARR280 pKa = 11.84NRR282 pKa = 11.84QSICRR287 pKa = 11.84SRR289 pKa = 11.84WEE291 pKa = 3.96PSFITLGVGQRR302 pKa = 11.84NAKK305 pKa = 10.36DD306 pKa = 3.23GAEE309 pKa = 3.94RR310 pKa = 3.8
Molecular weight: 34.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
922013 |
26 |
2401 |
316.7 |
35.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.532 ± 0.065 | 1.061 ± 0.019 |
5.13 ± 0.033 | 6.53 ± 0.047 |
3.575 ± 0.03 | 7.53 ± 0.051 |
2.092 ± 0.025 | 6.267 ± 0.04 |
4.759 ± 0.042 | 10.013 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.734 ± 0.021 | 3.348 ± 0.033 |
4.609 ± 0.038 | 4.56 ± 0.04 |
5.909 ± 0.037 | 5.195 ± 0.034 |
5.606 ± 0.034 | 7.392 ± 0.041 |
1.235 ± 0.018 | 2.922 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
