
Phytophthora parasitica (strain INRA-310)
Taxonomy: cellular organisms; Eukaryota; Sar; Stramenopiles; Oomycota; Peronosporales; Peronosporaceae; Phytophthora; Phytophthora parasitica
Average proteome isoelectric point is 6.64
Get precalculated fractions of proteins
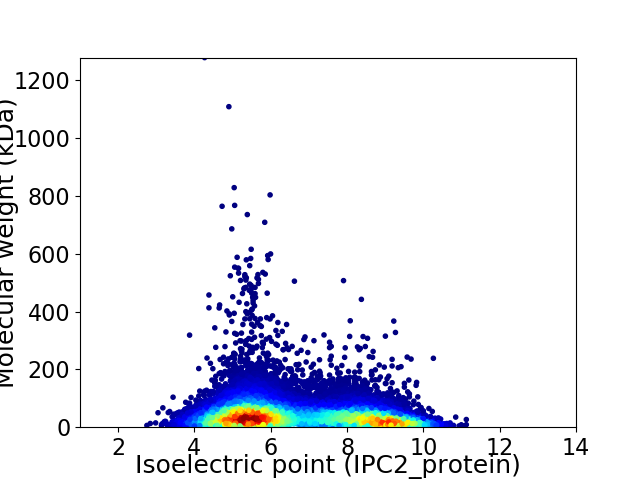
Virtual 2D-PAGE plot for 26438 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W2Q904|W2Q904_PHYPN FYVE-type domain-containing protein OS=Phytophthora parasitica (strain INRA-310) OX=761204 GN=PPTG_11262 PE=4 SV=1
MM1 pKa = 7.07QVFGVTALVFLALQGSVQAVQNAITCTTTADD32 pKa = 4.08CAWGEE37 pKa = 4.35TCVAGDD43 pKa = 4.38ADD45 pKa = 4.21TSVQACVAPTVCGGSSMGNCPSDD68 pKa = 3.53DD69 pKa = 3.83TGKK72 pKa = 10.26LACLWRR78 pKa = 11.84PFNDD82 pKa = 3.49CSEE85 pKa = 4.19GCAILNGNKK94 pKa = 10.11GIYY97 pKa = 9.36KK98 pKa = 9.7CVSITRR104 pKa = 11.84CDD106 pKa = 3.2AYY108 pKa = 11.34YY109 pKa = 10.86GGSKK113 pKa = 10.56CSDD116 pKa = 3.33GCSVNGVRR124 pKa = 11.84CNGQGSCNMMSSNADD139 pKa = 3.18GTPVFGCTCDD149 pKa = 3.06NGYY152 pKa = 10.63SGEE155 pKa = 4.16KK156 pKa = 10.23CEE158 pKa = 4.41NAPGSTNTSSTDD170 pKa = 3.12TDD172 pKa = 4.45DD173 pKa = 5.33EE174 pKa = 5.0DD175 pKa = 4.87SFWGALDD182 pKa = 5.35DD183 pKa = 5.92GSDD186 pKa = 4.48DD187 pKa = 4.07DD188 pKa = 4.86ASKK191 pKa = 11.23NGSDD195 pKa = 5.13DD196 pKa = 4.78ANADD200 pKa = 3.74DD201 pKa = 5.94SGTSDD206 pKa = 5.21DD207 pKa = 4.56KK208 pKa = 11.59STDD211 pKa = 3.3ASDD214 pKa = 5.54DD215 pKa = 3.73SATDD219 pKa = 3.38EE220 pKa = 4.69SSTDD224 pKa = 3.33TSDD227 pKa = 5.54DD228 pKa = 3.51SSTDD232 pKa = 3.34ADD234 pKa = 3.91QSTTDD239 pKa = 4.91ASDD242 pKa = 3.3EE243 pKa = 4.63SNADD247 pKa = 3.72DD248 pKa = 4.71NAEE251 pKa = 4.43DD252 pKa = 4.61DD253 pKa = 4.38SASSSSSASSSGEE266 pKa = 3.78LANSEE271 pKa = 4.76SVSSTSRR278 pKa = 11.84SGIRR282 pKa = 11.84PGVLILVLVMTAFFLVGTVLLVAYY306 pKa = 9.47SRR308 pKa = 11.84RR309 pKa = 11.84RR310 pKa = 11.84KK311 pKa = 7.81QQEE314 pKa = 3.7EE315 pKa = 4.23EE316 pKa = 4.47EE317 pKa = 4.48YY318 pKa = 11.43ANALASTQGVGAVGARR334 pKa = 11.84DD335 pKa = 3.4LAAGHH340 pKa = 6.21TGRR343 pKa = 11.84TPRR346 pKa = 11.84SNIARR351 pKa = 11.84MM352 pKa = 3.51
MM1 pKa = 7.07QVFGVTALVFLALQGSVQAVQNAITCTTTADD32 pKa = 4.08CAWGEE37 pKa = 4.35TCVAGDD43 pKa = 4.38ADD45 pKa = 4.21TSVQACVAPTVCGGSSMGNCPSDD68 pKa = 3.53DD69 pKa = 3.83TGKK72 pKa = 10.26LACLWRR78 pKa = 11.84PFNDD82 pKa = 3.49CSEE85 pKa = 4.19GCAILNGNKK94 pKa = 10.11GIYY97 pKa = 9.36KK98 pKa = 9.7CVSITRR104 pKa = 11.84CDD106 pKa = 3.2AYY108 pKa = 11.34YY109 pKa = 10.86GGSKK113 pKa = 10.56CSDD116 pKa = 3.33GCSVNGVRR124 pKa = 11.84CNGQGSCNMMSSNADD139 pKa = 3.18GTPVFGCTCDD149 pKa = 3.06NGYY152 pKa = 10.63SGEE155 pKa = 4.16KK156 pKa = 10.23CEE158 pKa = 4.41NAPGSTNTSSTDD170 pKa = 3.12TDD172 pKa = 4.45DD173 pKa = 5.33EE174 pKa = 5.0DD175 pKa = 4.87SFWGALDD182 pKa = 5.35DD183 pKa = 5.92GSDD186 pKa = 4.48DD187 pKa = 4.07DD188 pKa = 4.86ASKK191 pKa = 11.23NGSDD195 pKa = 5.13DD196 pKa = 4.78ANADD200 pKa = 3.74DD201 pKa = 5.94SGTSDD206 pKa = 5.21DD207 pKa = 4.56KK208 pKa = 11.59STDD211 pKa = 3.3ASDD214 pKa = 5.54DD215 pKa = 3.73SATDD219 pKa = 3.38EE220 pKa = 4.69SSTDD224 pKa = 3.33TSDD227 pKa = 5.54DD228 pKa = 3.51SSTDD232 pKa = 3.34ADD234 pKa = 3.91QSTTDD239 pKa = 4.91ASDD242 pKa = 3.3EE243 pKa = 4.63SNADD247 pKa = 3.72DD248 pKa = 4.71NAEE251 pKa = 4.43DD252 pKa = 4.61DD253 pKa = 4.38SASSSSSASSSGEE266 pKa = 3.78LANSEE271 pKa = 4.76SVSSTSRR278 pKa = 11.84SGIRR282 pKa = 11.84PGVLILVLVMTAFFLVGTVLLVAYY306 pKa = 9.47SRR308 pKa = 11.84RR309 pKa = 11.84RR310 pKa = 11.84KK311 pKa = 7.81QQEE314 pKa = 3.7EE315 pKa = 4.23EE316 pKa = 4.47EE317 pKa = 4.48YY318 pKa = 11.43ANALASTQGVGAVGARR334 pKa = 11.84DD335 pKa = 3.4LAAGHH340 pKa = 6.21TGRR343 pKa = 11.84TPRR346 pKa = 11.84SNIARR351 pKa = 11.84MM352 pKa = 3.51
Molecular weight: 35.96 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W2PB64|W2PB64_PHYPN Uncharacterized protein OS=Phytophthora parasitica (strain INRA-310) OX=761204 GN=PPTG_19898 PE=4 SV=1
PP1 pKa = 6.95RR2 pKa = 11.84TPRR5 pKa = 11.84PRR7 pKa = 11.84NLVIRR12 pKa = 11.84RR13 pKa = 11.84SLLRR17 pKa = 11.84SRR19 pKa = 11.84RR20 pKa = 11.84MSQLTLPPMHH30 pKa = 7.24PPMPLLRR37 pKa = 11.84RR38 pKa = 11.84LPTHH42 pKa = 6.83PLMHH46 pKa = 6.71LQMPPPMLQLTRR58 pKa = 11.84LPMRR62 pKa = 11.84QLMHH66 pKa = 7.36RR67 pKa = 11.84LMPQLTLLPMCRR79 pKa = 11.84PTRR82 pKa = 11.84RR83 pKa = 11.84QTSPPMPLLRR93 pKa = 11.84RR94 pKa = 11.84LPTHH98 pKa = 6.89PLMPPPMLQLTRR110 pKa = 11.84LPMRR114 pKa = 11.84QLMRR118 pKa = 11.84RR119 pKa = 11.84LMLQLTLLPMRR130 pKa = 11.84PPMRR134 pKa = 11.84LRR136 pKa = 11.84TRR138 pKa = 11.84PPMLLLRR145 pKa = 11.84RR146 pKa = 11.84LPTHH150 pKa = 6.89PLMPPPMLLLRR161 pKa = 11.84RR162 pKa = 11.84LPTHH166 pKa = 6.85PPMPPPMLQLTRR178 pKa = 11.84RR179 pKa = 11.84LILQLMCLPKK189 pKa = 10.4HH190 pKa = 6.06PLMLQLTRR198 pKa = 11.84PPMRR202 pKa = 11.84RR203 pKa = 11.84PTNLQHH209 pKa = 7.19PVTTKK214 pKa = 10.66VLTVLEE220 pKa = 4.37WRR222 pKa = 11.84TT223 pKa = 3.31
PP1 pKa = 6.95RR2 pKa = 11.84TPRR5 pKa = 11.84PRR7 pKa = 11.84NLVIRR12 pKa = 11.84RR13 pKa = 11.84SLLRR17 pKa = 11.84SRR19 pKa = 11.84RR20 pKa = 11.84MSQLTLPPMHH30 pKa = 7.24PPMPLLRR37 pKa = 11.84RR38 pKa = 11.84LPTHH42 pKa = 6.83PLMHH46 pKa = 6.71LQMPPPMLQLTRR58 pKa = 11.84LPMRR62 pKa = 11.84QLMHH66 pKa = 7.36RR67 pKa = 11.84LMPQLTLLPMCRR79 pKa = 11.84PTRR82 pKa = 11.84RR83 pKa = 11.84QTSPPMPLLRR93 pKa = 11.84RR94 pKa = 11.84LPTHH98 pKa = 6.89PLMPPPMLQLTRR110 pKa = 11.84LPMRR114 pKa = 11.84QLMRR118 pKa = 11.84RR119 pKa = 11.84LMLQLTLLPMRR130 pKa = 11.84PPMRR134 pKa = 11.84LRR136 pKa = 11.84TRR138 pKa = 11.84PPMLLLRR145 pKa = 11.84RR146 pKa = 11.84LPTHH150 pKa = 6.89PLMPPPMLLLRR161 pKa = 11.84RR162 pKa = 11.84LPTHH166 pKa = 6.85PPMPPPMLQLTRR178 pKa = 11.84RR179 pKa = 11.84LILQLMCLPKK189 pKa = 10.4HH190 pKa = 6.06PLMLQLTRR198 pKa = 11.84PPMRR202 pKa = 11.84RR203 pKa = 11.84PTNLQHH209 pKa = 7.19PVTTKK214 pKa = 10.66VLTVLEE220 pKa = 4.37WRR222 pKa = 11.84TT223 pKa = 3.31
Molecular weight: 26.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
10900847 |
30 |
11823 |
412.3 |
45.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.598 ± 0.014 | 1.653 ± 0.007 |
5.675 ± 0.011 | 6.695 ± 0.017 |
3.748 ± 0.01 | 5.86 ± 0.018 |
2.336 ± 0.007 | 4.264 ± 0.011 |
5.218 ± 0.018 | 9.389 ± 0.021 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.47 ± 0.007 | 3.678 ± 0.008 |
4.669 ± 0.015 | 4.262 ± 0.01 |
6.32 ± 0.016 | 8.354 ± 0.019 |
6.003 ± 0.014 | 7.006 ± 0.013 |
1.202 ± 0.005 | 2.6 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
