
Antarctic penguin virus B
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Avulavirinae; Orthoavulavirus; Avian orthoavulavirus 18
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins
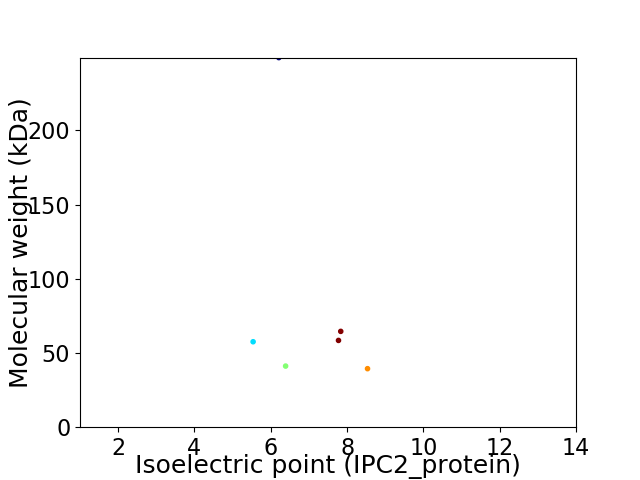
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y0KCB8|A0A1Y0KCB8_9MONO Nucleocapsid OS=Antarctic penguin virus B OX=2006073 PE=3 SV=1
MM1 pKa = 7.79ILRR4 pKa = 11.84IITGRR9 pKa = 11.84SRR11 pKa = 11.84YY12 pKa = 9.59SRR14 pKa = 11.84GRR16 pKa = 11.84DD17 pKa = 3.06RR18 pKa = 11.84EE19 pKa = 3.99QTYY22 pKa = 10.68SANHH26 pKa = 6.63SILNMSSIFSEE37 pKa = 3.82YY38 pKa = 11.04DD39 pKa = 2.93KK40 pKa = 11.19FLEE43 pKa = 4.74CQTHH47 pKa = 5.28PVSRR51 pKa = 11.84AMPTEE56 pKa = 4.19GGGALKK62 pKa = 10.31IEE64 pKa = 4.09IPVFVLNSDD73 pKa = 3.78DD74 pKa = 4.6PEE76 pKa = 4.25LRR78 pKa = 11.84WRR80 pKa = 11.84FTCFVLRR87 pKa = 11.84LALSEE92 pKa = 4.29SANRR96 pKa = 11.84PLRR99 pKa = 11.84QGALISVLCAHH110 pKa = 6.56AQVMKK115 pKa = 10.03TYY117 pKa = 10.81VSLAAQSGEE126 pKa = 3.84AVITILEE133 pKa = 4.12IDD135 pKa = 4.18DD136 pKa = 4.82FEE138 pKa = 6.31DD139 pKa = 4.88KK140 pKa = 11.02IPVFNNRR147 pKa = 11.84SGITEE152 pKa = 3.82EE153 pKa = 4.07RR154 pKa = 11.84ASRR157 pKa = 11.84LAMIAEE163 pKa = 4.52DD164 pKa = 4.8LPRR167 pKa = 11.84ACTNEE172 pKa = 4.05TPFVNGNTEE181 pKa = 3.88NDD183 pKa = 3.83PPEE186 pKa = 4.67DD187 pKa = 3.74ASDD190 pKa = 3.5VLDD193 pKa = 5.08RR194 pKa = 11.84IFSIQVQIWITVAKK208 pKa = 10.58AMTAFEE214 pKa = 4.14TAEE217 pKa = 3.87EE218 pKa = 4.62SEE220 pKa = 4.11TRR222 pKa = 11.84RR223 pKa = 11.84LNKK226 pKa = 9.94YY227 pKa = 6.6MQQGRR232 pKa = 11.84VQKK235 pKa = 10.47KK236 pKa = 9.16CLLFPIVRR244 pKa = 11.84TTVQLTIRR252 pKa = 11.84QSLLIRR258 pKa = 11.84GFLVSEE264 pKa = 4.61MKK266 pKa = 10.44RR267 pKa = 11.84AQNSPGGKK275 pKa = 7.92SAYY278 pKa = 9.76YY279 pKa = 10.66SFVGDD284 pKa = 3.68IAAYY288 pKa = 9.22IKK290 pKa = 10.76NAGVTAFLLTLKK302 pKa = 10.75FGIQTRR308 pKa = 11.84LPALALSSLSGDD320 pKa = 3.26IQKK323 pKa = 10.15VKK325 pKa = 10.84QLMVLYY331 pKa = 10.29RR332 pKa = 11.84EE333 pKa = 4.3KK334 pKa = 11.27GEE336 pKa = 4.07NGPYY340 pKa = 7.32MTLLGDD346 pKa = 3.91PDD348 pKa = 3.59QMQFAPAEE356 pKa = 4.01YY357 pKa = 10.29SLLYY361 pKa = 10.23SYY363 pKa = 11.85AMGVASVLEE372 pKa = 4.44SSTTRR377 pKa = 11.84YY378 pKa = 9.55QFARR382 pKa = 11.84DD383 pKa = 3.6FMNPTFWRR391 pKa = 11.84IGVEE395 pKa = 4.19SAQTLATSVDD405 pKa = 3.2EE406 pKa = 4.91GMASEE411 pKa = 4.4LQLGKK416 pKa = 9.94QSRR419 pKa = 11.84AVLSEE424 pKa = 3.63MMQKK428 pKa = 9.97VAGSAGEE435 pKa = 4.06YY436 pKa = 9.77TMNAPAASVMVGSGVGEE453 pKa = 4.09STRR456 pKa = 11.84PLTSKK461 pKa = 10.39VGSSRR466 pKa = 11.84IQSDD470 pKa = 4.13GAQIPPGFNTLEE482 pKa = 3.86EE483 pKa = 4.7YY484 pKa = 10.4YY485 pKa = 10.71DD486 pKa = 3.72YY487 pKa = 10.73MRR489 pKa = 11.84SEE491 pKa = 4.69GAGKK495 pKa = 10.36AGTSAGTSSKK505 pKa = 10.58PKK507 pKa = 10.3TPGLDD512 pKa = 4.09DD513 pKa = 4.35DD514 pKa = 5.08TDD516 pKa = 3.92NQGDD520 pKa = 3.39WSLL523 pKa = 3.62
MM1 pKa = 7.79ILRR4 pKa = 11.84IITGRR9 pKa = 11.84SRR11 pKa = 11.84YY12 pKa = 9.59SRR14 pKa = 11.84GRR16 pKa = 11.84DD17 pKa = 3.06RR18 pKa = 11.84EE19 pKa = 3.99QTYY22 pKa = 10.68SANHH26 pKa = 6.63SILNMSSIFSEE37 pKa = 3.82YY38 pKa = 11.04DD39 pKa = 2.93KK40 pKa = 11.19FLEE43 pKa = 4.74CQTHH47 pKa = 5.28PVSRR51 pKa = 11.84AMPTEE56 pKa = 4.19GGGALKK62 pKa = 10.31IEE64 pKa = 4.09IPVFVLNSDD73 pKa = 3.78DD74 pKa = 4.6PEE76 pKa = 4.25LRR78 pKa = 11.84WRR80 pKa = 11.84FTCFVLRR87 pKa = 11.84LALSEE92 pKa = 4.29SANRR96 pKa = 11.84PLRR99 pKa = 11.84QGALISVLCAHH110 pKa = 6.56AQVMKK115 pKa = 10.03TYY117 pKa = 10.81VSLAAQSGEE126 pKa = 3.84AVITILEE133 pKa = 4.12IDD135 pKa = 4.18DD136 pKa = 4.82FEE138 pKa = 6.31DD139 pKa = 4.88KK140 pKa = 11.02IPVFNNRR147 pKa = 11.84SGITEE152 pKa = 3.82EE153 pKa = 4.07RR154 pKa = 11.84ASRR157 pKa = 11.84LAMIAEE163 pKa = 4.52DD164 pKa = 4.8LPRR167 pKa = 11.84ACTNEE172 pKa = 4.05TPFVNGNTEE181 pKa = 3.88NDD183 pKa = 3.83PPEE186 pKa = 4.67DD187 pKa = 3.74ASDD190 pKa = 3.5VLDD193 pKa = 5.08RR194 pKa = 11.84IFSIQVQIWITVAKK208 pKa = 10.58AMTAFEE214 pKa = 4.14TAEE217 pKa = 3.87EE218 pKa = 4.62SEE220 pKa = 4.11TRR222 pKa = 11.84RR223 pKa = 11.84LNKK226 pKa = 9.94YY227 pKa = 6.6MQQGRR232 pKa = 11.84VQKK235 pKa = 10.47KK236 pKa = 9.16CLLFPIVRR244 pKa = 11.84TTVQLTIRR252 pKa = 11.84QSLLIRR258 pKa = 11.84GFLVSEE264 pKa = 4.61MKK266 pKa = 10.44RR267 pKa = 11.84AQNSPGGKK275 pKa = 7.92SAYY278 pKa = 9.76YY279 pKa = 10.66SFVGDD284 pKa = 3.68IAAYY288 pKa = 9.22IKK290 pKa = 10.76NAGVTAFLLTLKK302 pKa = 10.75FGIQTRR308 pKa = 11.84LPALALSSLSGDD320 pKa = 3.26IQKK323 pKa = 10.15VKK325 pKa = 10.84QLMVLYY331 pKa = 10.29RR332 pKa = 11.84EE333 pKa = 4.3KK334 pKa = 11.27GEE336 pKa = 4.07NGPYY340 pKa = 7.32MTLLGDD346 pKa = 3.91PDD348 pKa = 3.59QMQFAPAEE356 pKa = 4.01YY357 pKa = 10.29SLLYY361 pKa = 10.23SYY363 pKa = 11.85AMGVASVLEE372 pKa = 4.44SSTTRR377 pKa = 11.84YY378 pKa = 9.55QFARR382 pKa = 11.84DD383 pKa = 3.6FMNPTFWRR391 pKa = 11.84IGVEE395 pKa = 4.19SAQTLATSVDD405 pKa = 3.2EE406 pKa = 4.91GMASEE411 pKa = 4.4LQLGKK416 pKa = 9.94QSRR419 pKa = 11.84AVLSEE424 pKa = 3.63MMQKK428 pKa = 9.97VAGSAGEE435 pKa = 4.06YY436 pKa = 9.77TMNAPAASVMVGSGVGEE453 pKa = 4.09STRR456 pKa = 11.84PLTSKK461 pKa = 10.39VGSSRR466 pKa = 11.84IQSDD470 pKa = 4.13GAQIPPGFNTLEE482 pKa = 3.86EE483 pKa = 4.7YY484 pKa = 10.4YY485 pKa = 10.71DD486 pKa = 3.72YY487 pKa = 10.73MRR489 pKa = 11.84SEE491 pKa = 4.69GAGKK495 pKa = 10.36AGTSAGTSSKK505 pKa = 10.58PKK507 pKa = 10.3TPGLDD512 pKa = 4.09DD513 pKa = 4.35DD514 pKa = 5.08TDD516 pKa = 3.92NQGDD520 pKa = 3.39WSLL523 pKa = 3.62
Molecular weight: 57.57 kDa
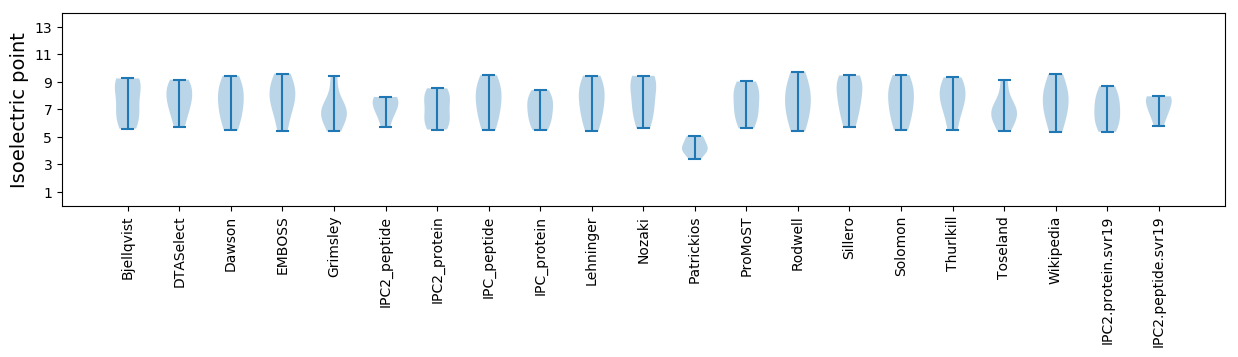
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y0KCB7|A0A1Y0KCB7_9MONO Phosphoprotein OS=Antarctic penguin virus B OX=2006073 PE=4 SV=1
MM1 pKa = 7.64SLEE4 pKa = 4.03SRR6 pKa = 11.84TVRR9 pKa = 11.84LYY11 pKa = 11.24VDD13 pKa = 4.12PDD15 pKa = 3.29QAASSLLAFPLIFTSNEE32 pKa = 3.97SGKK35 pKa = 8.68KK36 pKa = 7.8TLSPQYY42 pKa = 9.65RR43 pKa = 11.84IQIIEE48 pKa = 4.62DD49 pKa = 3.55DD50 pKa = 4.09RR51 pKa = 11.84EE52 pKa = 4.08SDD54 pKa = 3.31NDD56 pKa = 3.74LVFISTYY63 pKa = 10.99GFITGLEE70 pKa = 4.2TAVDD74 pKa = 3.9RR75 pKa = 11.84SISVDD80 pKa = 3.14MSQEE84 pKa = 3.95RR85 pKa = 11.84VVLTSCMLPLGSVPKK100 pKa = 10.47TSDD103 pKa = 2.9LHH105 pKa = 5.54EE106 pKa = 4.38LARR109 pKa = 11.84ACLEE113 pKa = 4.34LKK115 pKa = 9.8VACKK119 pKa = 10.03KK120 pKa = 10.53AATNSEE126 pKa = 4.1RR127 pKa = 11.84IIFNILDD134 pKa = 4.39APPVLAPCATIKK146 pKa = 10.63QAVTSCAAAVNLKK159 pKa = 10.54APEE162 pKa = 4.85KK163 pKa = 10.88IMGNYY168 pKa = 9.69DD169 pKa = 3.97LLYY172 pKa = 10.51KK173 pKa = 9.68VTFVSLTIIPAGSVYY188 pKa = 10.41KK189 pKa = 10.83VSSPVLKK196 pKa = 10.57AGSSLTYY203 pKa = 10.65GLNLSITLKK212 pKa = 10.28IDD214 pKa = 3.22IGDD217 pKa = 3.59QHH219 pKa = 7.05PSAKK223 pKa = 9.62MLMKK227 pKa = 10.27RR228 pKa = 11.84DD229 pKa = 3.54DD230 pKa = 3.74GFYY233 pKa = 10.46ANLWIHH239 pKa = 6.71CGLLSAVKK247 pKa = 10.39KK248 pKa = 10.44GGKK251 pKa = 8.34KK252 pKa = 9.94HH253 pKa = 6.38SIEE256 pKa = 4.29EE257 pKa = 3.89IANKK261 pKa = 8.9VRR263 pKa = 11.84RR264 pKa = 11.84LEE266 pKa = 4.08MKK268 pKa = 10.77LMLVDD273 pKa = 3.47MFGPSIVIKK282 pKa = 9.74CTGVKK287 pKa = 9.26TKK289 pKa = 10.64LLAGFFSKK297 pKa = 10.41KK298 pKa = 8.17GTAVYY303 pKa = 9.07PISRR307 pKa = 11.84AAPGIGKK314 pKa = 8.82LLWSQSGTITEE325 pKa = 4.0AAIIIQGGTPHH336 pKa = 6.28QLVSTSDD343 pKa = 3.65YY344 pKa = 11.16AVTSTKK350 pKa = 9.45VTVGGGNSKK359 pKa = 10.86YY360 pKa = 11.0NPFKK364 pKa = 10.83KK365 pKa = 10.46
MM1 pKa = 7.64SLEE4 pKa = 4.03SRR6 pKa = 11.84TVRR9 pKa = 11.84LYY11 pKa = 11.24VDD13 pKa = 4.12PDD15 pKa = 3.29QAASSLLAFPLIFTSNEE32 pKa = 3.97SGKK35 pKa = 8.68KK36 pKa = 7.8TLSPQYY42 pKa = 9.65RR43 pKa = 11.84IQIIEE48 pKa = 4.62DD49 pKa = 3.55DD50 pKa = 4.09RR51 pKa = 11.84EE52 pKa = 4.08SDD54 pKa = 3.31NDD56 pKa = 3.74LVFISTYY63 pKa = 10.99GFITGLEE70 pKa = 4.2TAVDD74 pKa = 3.9RR75 pKa = 11.84SISVDD80 pKa = 3.14MSQEE84 pKa = 3.95RR85 pKa = 11.84VVLTSCMLPLGSVPKK100 pKa = 10.47TSDD103 pKa = 2.9LHH105 pKa = 5.54EE106 pKa = 4.38LARR109 pKa = 11.84ACLEE113 pKa = 4.34LKK115 pKa = 9.8VACKK119 pKa = 10.03KK120 pKa = 10.53AATNSEE126 pKa = 4.1RR127 pKa = 11.84IIFNILDD134 pKa = 4.39APPVLAPCATIKK146 pKa = 10.63QAVTSCAAAVNLKK159 pKa = 10.54APEE162 pKa = 4.85KK163 pKa = 10.88IMGNYY168 pKa = 9.69DD169 pKa = 3.97LLYY172 pKa = 10.51KK173 pKa = 9.68VTFVSLTIIPAGSVYY188 pKa = 10.41KK189 pKa = 10.83VSSPVLKK196 pKa = 10.57AGSSLTYY203 pKa = 10.65GLNLSITLKK212 pKa = 10.28IDD214 pKa = 3.22IGDD217 pKa = 3.59QHH219 pKa = 7.05PSAKK223 pKa = 9.62MLMKK227 pKa = 10.27RR228 pKa = 11.84DD229 pKa = 3.54DD230 pKa = 3.74GFYY233 pKa = 10.46ANLWIHH239 pKa = 6.71CGLLSAVKK247 pKa = 10.39KK248 pKa = 10.44GGKK251 pKa = 8.34KK252 pKa = 9.94HH253 pKa = 6.38SIEE256 pKa = 4.29EE257 pKa = 3.89IANKK261 pKa = 8.9VRR263 pKa = 11.84RR264 pKa = 11.84LEE266 pKa = 4.08MKK268 pKa = 10.77LMLVDD273 pKa = 3.47MFGPSIVIKK282 pKa = 9.74CTGVKK287 pKa = 9.26TKK289 pKa = 10.64LLAGFFSKK297 pKa = 10.41KK298 pKa = 8.17GTAVYY303 pKa = 9.07PISRR307 pKa = 11.84AAPGIGKK314 pKa = 8.82LLWSQSGTITEE325 pKa = 4.0AAIIIQGGTPHH336 pKa = 6.28QLVSTSDD343 pKa = 3.65YY344 pKa = 11.16AVTSTKK350 pKa = 9.45VTVGGGNSKK359 pKa = 10.86YY360 pKa = 11.0NPFKK364 pKa = 10.83KK365 pKa = 10.46
Molecular weight: 39.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4621 |
365 |
2207 |
770.2 |
85.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.531 ± 0.536 | 2.056 ± 0.239 |
4.891 ± 0.477 | 4.696 ± 0.421 |
3.073 ± 0.346 | 5.865 ± 0.634 |
1.536 ± 0.413 | 6.709 ± 0.514 |
4.761 ± 0.448 | 10.777 ± 1.091 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.792 ± 0.154 | 4.588 ± 0.333 |
4.804 ± 0.538 | 4.458 ± 0.415 |
5.129 ± 0.536 | 9.046 ± 0.597 |
6.557 ± 0.39 | 6.47 ± 0.621 |
0.844 ± 0.151 | 3.419 ± 0.388 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
