
Beihai weivirus-like virus 18
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 5.8
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KLB4|A0A1L3KLB4_9VIRU RNA-dependent RNA polymerase OS=Beihai weivirus-like virus 18 OX=1922746 PE=4 SV=1
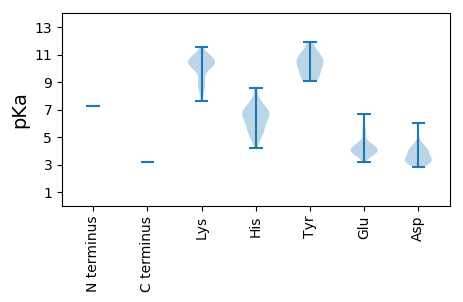
MM1 pKa = 7.24FGEE4 pKa = 4.65SKK6 pKa = 10.55FGSTFPEE13 pKa = 3.94ICEE16 pKa = 4.09TCHH19 pKa = 7.47RR20 pKa = 11.84DD21 pKa = 3.26EE22 pKa = 5.15LQFLSKK28 pKa = 10.17PRR30 pKa = 11.84SSEE33 pKa = 3.73DD34 pKa = 2.81HH35 pKa = 5.22MTRR38 pKa = 11.84VMTIKK43 pKa = 10.46RR44 pKa = 11.84CCTRR48 pKa = 11.84CNNTCFQWPEE58 pKa = 3.53FLGYY62 pKa = 10.45CDD64 pKa = 3.48VTICLPVSLVADD76 pKa = 3.71STSFAQRR83 pKa = 11.84HH84 pKa = 4.17QASTKK89 pKa = 8.88WGIKK93 pKa = 9.17PSCADD98 pKa = 3.42LQLVNLEE105 pKa = 4.21CAARR109 pKa = 11.84DD110 pKa = 3.58PFRR113 pKa = 11.84RR114 pKa = 11.84RR115 pKa = 11.84AYY117 pKa = 9.57LTVGHH122 pKa = 7.05AAMLDD127 pKa = 3.36EE128 pKa = 5.39LVFHH132 pKa = 7.04GPGLLDD138 pKa = 6.04CILQLCCFRR147 pKa = 11.84NAKK150 pKa = 10.28GSTSWRR156 pKa = 11.84LCNSARR162 pKa = 11.84SNQSEE167 pKa = 4.31PPLDD171 pKa = 4.82RR172 pKa = 11.84IQEE175 pKa = 3.99VDD177 pKa = 4.03EE178 pKa = 5.68GDD180 pKa = 4.39DD181 pKa = 3.9NQGDD185 pKa = 4.32DD186 pKa = 4.06VNPAAAHH193 pKa = 5.37SPSRR197 pKa = 11.84NANHH201 pKa = 5.35QQQNGGRR208 pKa = 11.84TEE210 pKa = 4.26AEE212 pKa = 3.99LALEE216 pKa = 4.15QALEE220 pKa = 3.96ASNVEE225 pKa = 4.24EE226 pKa = 3.76QRR228 pKa = 11.84RR229 pKa = 11.84AYY231 pKa = 10.77VEE233 pKa = 4.09GGVLHH238 pKa = 6.19STTTLAPGQRR248 pKa = 11.84PEE250 pKa = 4.43LVEE253 pKa = 4.02PPGLEE258 pKa = 4.22GGCASGTRR266 pKa = 11.84QARR269 pKa = 11.84ARR271 pKa = 11.84FPQITSQPNFLHH283 pKa = 7.0SNNEE287 pKa = 3.66QNLQMAHH294 pKa = 6.77DD295 pKa = 4.05MRR297 pKa = 11.84NVGVGDD303 pKa = 4.16HH304 pKa = 6.83NPLQSEE310 pKa = 4.47VQTRR314 pKa = 11.84DD315 pKa = 3.17RR316 pKa = 11.84LVEE319 pKa = 3.85LLKK322 pKa = 10.9QKK324 pKa = 10.38VFTPGRR330 pKa = 11.84LRR332 pKa = 11.84KK333 pKa = 10.14AMVGFEE339 pKa = 4.23SARR342 pKa = 11.84VSALPKK348 pKa = 10.18KK349 pKa = 10.43LSEE352 pKa = 4.22EE353 pKa = 3.42EE354 pKa = 3.91RR355 pKa = 11.84MRR357 pKa = 11.84VEE359 pKa = 4.24LEE361 pKa = 3.83AMNAALSDD369 pKa = 4.06DD370 pKa = 3.77GVGFDD375 pKa = 3.77TVIKK379 pKa = 10.49AFVKK383 pKa = 10.84SEE385 pKa = 3.74VTAKK389 pKa = 10.63NKK391 pKa = 8.89PRR393 pKa = 11.84PIANHH398 pKa = 5.46GNVRR402 pKa = 11.84LYY404 pKa = 11.29ALAKK408 pKa = 8.44VAYY411 pKa = 9.07AFEE414 pKa = 4.46HH415 pKa = 6.51VMFEE419 pKa = 4.3TFKK422 pKa = 11.07GGSIKK427 pKa = 10.68GRR429 pKa = 11.84GKK431 pKa = 10.39KK432 pKa = 8.81EE433 pKa = 3.96AIEE436 pKa = 3.96EE437 pKa = 4.3LFSNMSNMKK446 pKa = 9.73NGRR449 pKa = 11.84WVEE452 pKa = 3.72NDD454 pKa = 2.97LTSFEE459 pKa = 4.53FGISEE464 pKa = 4.16PLKK467 pKa = 10.27QIEE470 pKa = 3.93MDD472 pKa = 3.34IFYY475 pKa = 9.97HH476 pKa = 5.46IAKK479 pKa = 9.84IIGVADD485 pKa = 3.98TGALLFEE492 pKa = 4.65RR493 pKa = 11.84VSNDD497 pKa = 3.12RR498 pKa = 11.84DD499 pKa = 3.4KK500 pKa = 11.55CATWKK505 pKa = 10.68LIYY508 pKa = 10.1TDD510 pKa = 3.15EE511 pKa = 4.5TGQKK515 pKa = 8.38RR516 pKa = 11.84TAKK519 pKa = 10.08IKK521 pKa = 10.17IPQTMRR527 pKa = 11.84EE528 pKa = 3.95SGDD531 pKa = 3.37RR532 pKa = 11.84VTSSGNFLQNLIAWFCFLVDD552 pKa = 3.56PDD554 pKa = 4.08YY555 pKa = 11.88VEE557 pKa = 5.48DD558 pKa = 4.34ALEE561 pKa = 3.94SLIKK565 pKa = 10.6HH566 pKa = 6.21RR567 pKa = 11.84GSKK570 pKa = 10.18LFYY573 pKa = 9.38VSRR576 pKa = 11.84RR577 pKa = 11.84DD578 pKa = 3.09RR579 pKa = 11.84STIEE583 pKa = 3.82LRR585 pKa = 11.84GKK587 pKa = 7.62QVRR590 pKa = 11.84KK591 pKa = 10.19KK592 pKa = 10.75YY593 pKa = 9.56LACLAFEE600 pKa = 5.22GDD602 pKa = 3.83DD603 pKa = 3.25TSGRR607 pKa = 11.84IEE609 pKa = 4.05EE610 pKa = 4.41PVWLNGPYY618 pKa = 10.55NPDD621 pKa = 3.14NSEE624 pKa = 4.06TKK626 pKa = 10.46DD627 pKa = 3.46DD628 pKa = 4.27CLVSSFFRR636 pKa = 11.84RR637 pKa = 11.84WGWKK641 pKa = 9.75AKK643 pKa = 10.3LIWKK647 pKa = 8.09PQEE650 pKa = 3.21GDD652 pKa = 3.16AYY654 pKa = 10.87VRR656 pKa = 11.84FVGYY660 pKa = 9.12EE661 pKa = 3.87ALIHH665 pKa = 6.61DD666 pKa = 4.62SKK668 pKa = 10.95IVYY671 pKa = 10.12DD672 pKa = 3.85GAEE675 pKa = 3.66MVMTPEE681 pKa = 4.71ARR683 pKa = 11.84RR684 pKa = 11.84LLNTKK689 pKa = 8.93SWTTTAVTPAEE700 pKa = 4.52LKK702 pKa = 8.98TCIRR706 pKa = 11.84VYY708 pKa = 10.5AASLADD714 pKa = 3.31GFKK717 pKa = 10.42RR718 pKa = 11.84VEE720 pKa = 3.73PMYY723 pKa = 11.2AFLNAVFDD731 pKa = 4.44SNKK734 pKa = 10.52GGVDD738 pKa = 3.09VDD740 pKa = 3.62AQKK743 pKa = 11.11VKK745 pKa = 10.58EE746 pKa = 3.98YY747 pKa = 10.6LLAVEE752 pKa = 5.45GEE754 pKa = 4.32MPEE757 pKa = 3.73HH758 pKa = 6.63GRR760 pKa = 11.84TVNCPIDD767 pKa = 3.57MPPFEE772 pKa = 6.7GGDD775 pKa = 3.22ADD777 pKa = 3.05KK778 pKa = 10.65WKK780 pKa = 10.67RR781 pKa = 11.84LLRR784 pKa = 11.84RR785 pKa = 11.84SAGDD789 pKa = 3.12FDD791 pKa = 3.75EE792 pKa = 6.18RR793 pKa = 11.84EE794 pKa = 4.14WATMCHH800 pKa = 6.57ISTIDD805 pKa = 3.06IHH807 pKa = 8.6GEE809 pKa = 3.64DD810 pKa = 4.16LAISVPAEE818 pKa = 3.59WRR820 pKa = 11.84NN821 pKa = 3.21
MM1 pKa = 7.24FGEE4 pKa = 4.65SKK6 pKa = 10.55FGSTFPEE13 pKa = 3.94ICEE16 pKa = 4.09TCHH19 pKa = 7.47RR20 pKa = 11.84DD21 pKa = 3.26EE22 pKa = 5.15LQFLSKK28 pKa = 10.17PRR30 pKa = 11.84SSEE33 pKa = 3.73DD34 pKa = 2.81HH35 pKa = 5.22MTRR38 pKa = 11.84VMTIKK43 pKa = 10.46RR44 pKa = 11.84CCTRR48 pKa = 11.84CNNTCFQWPEE58 pKa = 3.53FLGYY62 pKa = 10.45CDD64 pKa = 3.48VTICLPVSLVADD76 pKa = 3.71STSFAQRR83 pKa = 11.84HH84 pKa = 4.17QASTKK89 pKa = 8.88WGIKK93 pKa = 9.17PSCADD98 pKa = 3.42LQLVNLEE105 pKa = 4.21CAARR109 pKa = 11.84DD110 pKa = 3.58PFRR113 pKa = 11.84RR114 pKa = 11.84RR115 pKa = 11.84AYY117 pKa = 9.57LTVGHH122 pKa = 7.05AAMLDD127 pKa = 3.36EE128 pKa = 5.39LVFHH132 pKa = 7.04GPGLLDD138 pKa = 6.04CILQLCCFRR147 pKa = 11.84NAKK150 pKa = 10.28GSTSWRR156 pKa = 11.84LCNSARR162 pKa = 11.84SNQSEE167 pKa = 4.31PPLDD171 pKa = 4.82RR172 pKa = 11.84IQEE175 pKa = 3.99VDD177 pKa = 4.03EE178 pKa = 5.68GDD180 pKa = 4.39DD181 pKa = 3.9NQGDD185 pKa = 4.32DD186 pKa = 4.06VNPAAAHH193 pKa = 5.37SPSRR197 pKa = 11.84NANHH201 pKa = 5.35QQQNGGRR208 pKa = 11.84TEE210 pKa = 4.26AEE212 pKa = 3.99LALEE216 pKa = 4.15QALEE220 pKa = 3.96ASNVEE225 pKa = 4.24EE226 pKa = 3.76QRR228 pKa = 11.84RR229 pKa = 11.84AYY231 pKa = 10.77VEE233 pKa = 4.09GGVLHH238 pKa = 6.19STTTLAPGQRR248 pKa = 11.84PEE250 pKa = 4.43LVEE253 pKa = 4.02PPGLEE258 pKa = 4.22GGCASGTRR266 pKa = 11.84QARR269 pKa = 11.84ARR271 pKa = 11.84FPQITSQPNFLHH283 pKa = 7.0SNNEE287 pKa = 3.66QNLQMAHH294 pKa = 6.77DD295 pKa = 4.05MRR297 pKa = 11.84NVGVGDD303 pKa = 4.16HH304 pKa = 6.83NPLQSEE310 pKa = 4.47VQTRR314 pKa = 11.84DD315 pKa = 3.17RR316 pKa = 11.84LVEE319 pKa = 3.85LLKK322 pKa = 10.9QKK324 pKa = 10.38VFTPGRR330 pKa = 11.84LRR332 pKa = 11.84KK333 pKa = 10.14AMVGFEE339 pKa = 4.23SARR342 pKa = 11.84VSALPKK348 pKa = 10.18KK349 pKa = 10.43LSEE352 pKa = 4.22EE353 pKa = 3.42EE354 pKa = 3.91RR355 pKa = 11.84MRR357 pKa = 11.84VEE359 pKa = 4.24LEE361 pKa = 3.83AMNAALSDD369 pKa = 4.06DD370 pKa = 3.77GVGFDD375 pKa = 3.77TVIKK379 pKa = 10.49AFVKK383 pKa = 10.84SEE385 pKa = 3.74VTAKK389 pKa = 10.63NKK391 pKa = 8.89PRR393 pKa = 11.84PIANHH398 pKa = 5.46GNVRR402 pKa = 11.84LYY404 pKa = 11.29ALAKK408 pKa = 8.44VAYY411 pKa = 9.07AFEE414 pKa = 4.46HH415 pKa = 6.51VMFEE419 pKa = 4.3TFKK422 pKa = 11.07GGSIKK427 pKa = 10.68GRR429 pKa = 11.84GKK431 pKa = 10.39KK432 pKa = 8.81EE433 pKa = 3.96AIEE436 pKa = 3.96EE437 pKa = 4.3LFSNMSNMKK446 pKa = 9.73NGRR449 pKa = 11.84WVEE452 pKa = 3.72NDD454 pKa = 2.97LTSFEE459 pKa = 4.53FGISEE464 pKa = 4.16PLKK467 pKa = 10.27QIEE470 pKa = 3.93MDD472 pKa = 3.34IFYY475 pKa = 9.97HH476 pKa = 5.46IAKK479 pKa = 9.84IIGVADD485 pKa = 3.98TGALLFEE492 pKa = 4.65RR493 pKa = 11.84VSNDD497 pKa = 3.12RR498 pKa = 11.84DD499 pKa = 3.4KK500 pKa = 11.55CATWKK505 pKa = 10.68LIYY508 pKa = 10.1TDD510 pKa = 3.15EE511 pKa = 4.5TGQKK515 pKa = 8.38RR516 pKa = 11.84TAKK519 pKa = 10.08IKK521 pKa = 10.17IPQTMRR527 pKa = 11.84EE528 pKa = 3.95SGDD531 pKa = 3.37RR532 pKa = 11.84VTSSGNFLQNLIAWFCFLVDD552 pKa = 3.56PDD554 pKa = 4.08YY555 pKa = 11.88VEE557 pKa = 5.48DD558 pKa = 4.34ALEE561 pKa = 3.94SLIKK565 pKa = 10.6HH566 pKa = 6.21RR567 pKa = 11.84GSKK570 pKa = 10.18LFYY573 pKa = 9.38VSRR576 pKa = 11.84RR577 pKa = 11.84DD578 pKa = 3.09RR579 pKa = 11.84STIEE583 pKa = 3.82LRR585 pKa = 11.84GKK587 pKa = 7.62QVRR590 pKa = 11.84KK591 pKa = 10.19KK592 pKa = 10.75YY593 pKa = 9.56LACLAFEE600 pKa = 5.22GDD602 pKa = 3.83DD603 pKa = 3.25TSGRR607 pKa = 11.84IEE609 pKa = 4.05EE610 pKa = 4.41PVWLNGPYY618 pKa = 10.55NPDD621 pKa = 3.14NSEE624 pKa = 4.06TKK626 pKa = 10.46DD627 pKa = 3.46DD628 pKa = 4.27CLVSSFFRR636 pKa = 11.84RR637 pKa = 11.84WGWKK641 pKa = 9.75AKK643 pKa = 10.3LIWKK647 pKa = 8.09PQEE650 pKa = 3.21GDD652 pKa = 3.16AYY654 pKa = 10.87VRR656 pKa = 11.84FVGYY660 pKa = 9.12EE661 pKa = 3.87ALIHH665 pKa = 6.61DD666 pKa = 4.62SKK668 pKa = 10.95IVYY671 pKa = 10.12DD672 pKa = 3.85GAEE675 pKa = 3.66MVMTPEE681 pKa = 4.71ARR683 pKa = 11.84RR684 pKa = 11.84LLNTKK689 pKa = 8.93SWTTTAVTPAEE700 pKa = 4.52LKK702 pKa = 8.98TCIRR706 pKa = 11.84VYY708 pKa = 10.5AASLADD714 pKa = 3.31GFKK717 pKa = 10.42RR718 pKa = 11.84VEE720 pKa = 3.73PMYY723 pKa = 11.2AFLNAVFDD731 pKa = 4.44SNKK734 pKa = 10.52GGVDD738 pKa = 3.09VDD740 pKa = 3.62AQKK743 pKa = 11.11VKK745 pKa = 10.58EE746 pKa = 3.98YY747 pKa = 10.6LLAVEE752 pKa = 5.45GEE754 pKa = 4.32MPEE757 pKa = 3.73HH758 pKa = 6.63GRR760 pKa = 11.84TVNCPIDD767 pKa = 3.57MPPFEE772 pKa = 6.7GGDD775 pKa = 3.22ADD777 pKa = 3.05KK778 pKa = 10.65WKK780 pKa = 10.67RR781 pKa = 11.84LLRR784 pKa = 11.84RR785 pKa = 11.84SAGDD789 pKa = 3.12FDD791 pKa = 3.75EE792 pKa = 6.18RR793 pKa = 11.84EE794 pKa = 4.14WATMCHH800 pKa = 6.57ISTIDD805 pKa = 3.06IHH807 pKa = 8.6GEE809 pKa = 3.64DD810 pKa = 4.16LAISVPAEE818 pKa = 3.59WRR820 pKa = 11.84NN821 pKa = 3.21
Molecular weight: 92.39 kDa
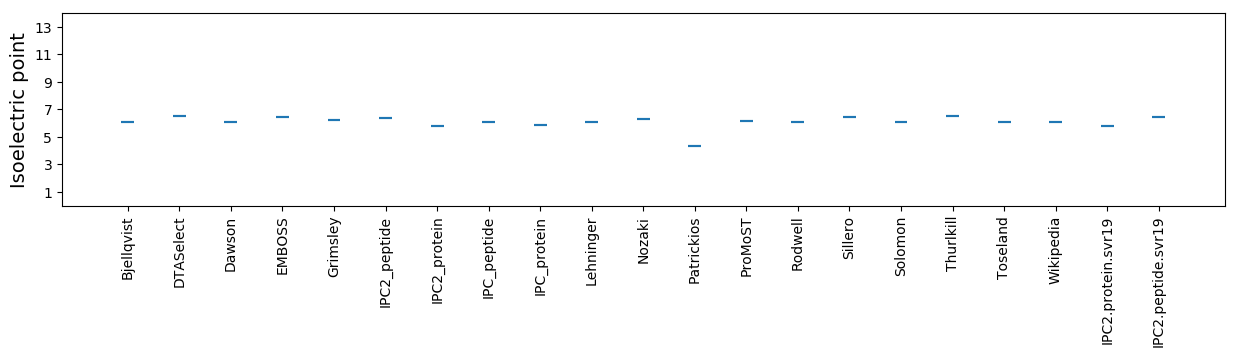
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KLB4|A0A1L3KLB4_9VIRU RNA-dependent RNA polymerase OS=Beihai weivirus-like virus 18 OX=1922746 PE=4 SV=1
MM1 pKa = 7.24FGEE4 pKa = 4.65SKK6 pKa = 10.55FGSTFPEE13 pKa = 3.94ICEE16 pKa = 4.09TCHH19 pKa = 7.47RR20 pKa = 11.84DD21 pKa = 3.26EE22 pKa = 5.15LQFLSKK28 pKa = 10.17PRR30 pKa = 11.84SSEE33 pKa = 3.73DD34 pKa = 2.81HH35 pKa = 5.22MTRR38 pKa = 11.84VMTIKK43 pKa = 10.46RR44 pKa = 11.84CCTRR48 pKa = 11.84CNNTCFQWPEE58 pKa = 3.53FLGYY62 pKa = 10.45CDD64 pKa = 3.48VTICLPVSLVADD76 pKa = 3.71STSFAQRR83 pKa = 11.84HH84 pKa = 4.17QASTKK89 pKa = 8.88WGIKK93 pKa = 9.17PSCADD98 pKa = 3.42LQLVNLEE105 pKa = 4.21CAARR109 pKa = 11.84DD110 pKa = 3.58PFRR113 pKa = 11.84RR114 pKa = 11.84RR115 pKa = 11.84AYY117 pKa = 9.57LTVGHH122 pKa = 7.05AAMLDD127 pKa = 3.36EE128 pKa = 5.39LVFHH132 pKa = 7.04GPGLLDD138 pKa = 6.04CILQLCCFRR147 pKa = 11.84NAKK150 pKa = 10.28GSTSWRR156 pKa = 11.84LCNSARR162 pKa = 11.84SNQSEE167 pKa = 4.31PPLDD171 pKa = 4.82RR172 pKa = 11.84IQEE175 pKa = 3.99VDD177 pKa = 4.03EE178 pKa = 5.68GDD180 pKa = 4.39DD181 pKa = 3.9NQGDD185 pKa = 4.32DD186 pKa = 4.06VNPAAAHH193 pKa = 5.37SPSRR197 pKa = 11.84NANHH201 pKa = 5.35QQQNGGRR208 pKa = 11.84TEE210 pKa = 4.26AEE212 pKa = 3.99LALEE216 pKa = 4.15QALEE220 pKa = 3.96ASNVEE225 pKa = 4.24EE226 pKa = 3.76QRR228 pKa = 11.84RR229 pKa = 11.84AYY231 pKa = 10.77VEE233 pKa = 4.09GGVLHH238 pKa = 6.19STTTLAPGQRR248 pKa = 11.84PEE250 pKa = 4.43LVEE253 pKa = 4.02PPGLEE258 pKa = 4.22GGCASGTRR266 pKa = 11.84QARR269 pKa = 11.84ARR271 pKa = 11.84FPQITSQPNFLHH283 pKa = 7.0SNNEE287 pKa = 3.66QNLQMAHH294 pKa = 6.77DD295 pKa = 4.05MRR297 pKa = 11.84NVGVGDD303 pKa = 4.16HH304 pKa = 6.83NPLQSEE310 pKa = 4.47VQTRR314 pKa = 11.84DD315 pKa = 3.17RR316 pKa = 11.84LVEE319 pKa = 3.85LLKK322 pKa = 10.9QKK324 pKa = 10.38VFTPGRR330 pKa = 11.84LRR332 pKa = 11.84KK333 pKa = 10.14AMVGFEE339 pKa = 4.23SARR342 pKa = 11.84VSALPKK348 pKa = 10.18KK349 pKa = 10.43LSEE352 pKa = 4.22EE353 pKa = 3.42EE354 pKa = 3.91RR355 pKa = 11.84MRR357 pKa = 11.84VEE359 pKa = 4.24LEE361 pKa = 3.83AMNAALSDD369 pKa = 4.06DD370 pKa = 3.77GVGFDD375 pKa = 3.77TVIKK379 pKa = 10.49AFVKK383 pKa = 10.84SEE385 pKa = 3.74VTAKK389 pKa = 10.63NKK391 pKa = 8.89PRR393 pKa = 11.84PIANHH398 pKa = 5.46GNVRR402 pKa = 11.84LYY404 pKa = 11.29ALAKK408 pKa = 8.44VAYY411 pKa = 9.07AFEE414 pKa = 4.46HH415 pKa = 6.51VMFEE419 pKa = 4.3TFKK422 pKa = 11.07GGSIKK427 pKa = 10.68GRR429 pKa = 11.84GKK431 pKa = 10.39KK432 pKa = 8.81EE433 pKa = 3.96AIEE436 pKa = 3.96EE437 pKa = 4.3LFSNMSNMKK446 pKa = 9.73NGRR449 pKa = 11.84WVEE452 pKa = 3.72NDD454 pKa = 2.97LTSFEE459 pKa = 4.53FGISEE464 pKa = 4.16PLKK467 pKa = 10.27QIEE470 pKa = 3.93MDD472 pKa = 3.34IFYY475 pKa = 9.97HH476 pKa = 5.46IAKK479 pKa = 9.84IIGVADD485 pKa = 3.98TGALLFEE492 pKa = 4.65RR493 pKa = 11.84VSNDD497 pKa = 3.12RR498 pKa = 11.84DD499 pKa = 3.4KK500 pKa = 11.55CATWKK505 pKa = 10.68LIYY508 pKa = 10.1TDD510 pKa = 3.15EE511 pKa = 4.5TGQKK515 pKa = 8.38RR516 pKa = 11.84TAKK519 pKa = 10.08IKK521 pKa = 10.17IPQTMRR527 pKa = 11.84EE528 pKa = 3.95SGDD531 pKa = 3.37RR532 pKa = 11.84VTSSGNFLQNLIAWFCFLVDD552 pKa = 3.56PDD554 pKa = 4.08YY555 pKa = 11.88VEE557 pKa = 5.48DD558 pKa = 4.34ALEE561 pKa = 3.94SLIKK565 pKa = 10.6HH566 pKa = 6.21RR567 pKa = 11.84GSKK570 pKa = 10.18LFYY573 pKa = 9.38VSRR576 pKa = 11.84RR577 pKa = 11.84DD578 pKa = 3.09RR579 pKa = 11.84STIEE583 pKa = 3.82LRR585 pKa = 11.84GKK587 pKa = 7.62QVRR590 pKa = 11.84KK591 pKa = 10.19KK592 pKa = 10.75YY593 pKa = 9.56LACLAFEE600 pKa = 5.22GDD602 pKa = 3.83DD603 pKa = 3.25TSGRR607 pKa = 11.84IEE609 pKa = 4.05EE610 pKa = 4.41PVWLNGPYY618 pKa = 10.55NPDD621 pKa = 3.14NSEE624 pKa = 4.06TKK626 pKa = 10.46DD627 pKa = 3.46DD628 pKa = 4.27CLVSSFFRR636 pKa = 11.84RR637 pKa = 11.84WGWKK641 pKa = 9.75AKK643 pKa = 10.3LIWKK647 pKa = 8.09PQEE650 pKa = 3.21GDD652 pKa = 3.16AYY654 pKa = 10.87VRR656 pKa = 11.84FVGYY660 pKa = 9.12EE661 pKa = 3.87ALIHH665 pKa = 6.61DD666 pKa = 4.62SKK668 pKa = 10.95IVYY671 pKa = 10.12DD672 pKa = 3.85GAEE675 pKa = 3.66MVMTPEE681 pKa = 4.71ARR683 pKa = 11.84RR684 pKa = 11.84LLNTKK689 pKa = 8.93SWTTTAVTPAEE700 pKa = 4.52LKK702 pKa = 8.98TCIRR706 pKa = 11.84VYY708 pKa = 10.5AASLADD714 pKa = 3.31GFKK717 pKa = 10.42RR718 pKa = 11.84VEE720 pKa = 3.73PMYY723 pKa = 11.2AFLNAVFDD731 pKa = 4.44SNKK734 pKa = 10.52GGVDD738 pKa = 3.09VDD740 pKa = 3.62AQKK743 pKa = 11.11VKK745 pKa = 10.58EE746 pKa = 3.98YY747 pKa = 10.6LLAVEE752 pKa = 5.45GEE754 pKa = 4.32MPEE757 pKa = 3.73HH758 pKa = 6.63GRR760 pKa = 11.84TVNCPIDD767 pKa = 3.57MPPFEE772 pKa = 6.7GGDD775 pKa = 3.22ADD777 pKa = 3.05KK778 pKa = 10.65WKK780 pKa = 10.67RR781 pKa = 11.84LLRR784 pKa = 11.84RR785 pKa = 11.84SAGDD789 pKa = 3.12FDD791 pKa = 3.75EE792 pKa = 6.18RR793 pKa = 11.84EE794 pKa = 4.14WATMCHH800 pKa = 6.57ISTIDD805 pKa = 3.06IHH807 pKa = 8.6GEE809 pKa = 3.64DD810 pKa = 4.16LAISVPAEE818 pKa = 3.59WRR820 pKa = 11.84NN821 pKa = 3.21
MM1 pKa = 7.24FGEE4 pKa = 4.65SKK6 pKa = 10.55FGSTFPEE13 pKa = 3.94ICEE16 pKa = 4.09TCHH19 pKa = 7.47RR20 pKa = 11.84DD21 pKa = 3.26EE22 pKa = 5.15LQFLSKK28 pKa = 10.17PRR30 pKa = 11.84SSEE33 pKa = 3.73DD34 pKa = 2.81HH35 pKa = 5.22MTRR38 pKa = 11.84VMTIKK43 pKa = 10.46RR44 pKa = 11.84CCTRR48 pKa = 11.84CNNTCFQWPEE58 pKa = 3.53FLGYY62 pKa = 10.45CDD64 pKa = 3.48VTICLPVSLVADD76 pKa = 3.71STSFAQRR83 pKa = 11.84HH84 pKa = 4.17QASTKK89 pKa = 8.88WGIKK93 pKa = 9.17PSCADD98 pKa = 3.42LQLVNLEE105 pKa = 4.21CAARR109 pKa = 11.84DD110 pKa = 3.58PFRR113 pKa = 11.84RR114 pKa = 11.84RR115 pKa = 11.84AYY117 pKa = 9.57LTVGHH122 pKa = 7.05AAMLDD127 pKa = 3.36EE128 pKa = 5.39LVFHH132 pKa = 7.04GPGLLDD138 pKa = 6.04CILQLCCFRR147 pKa = 11.84NAKK150 pKa = 10.28GSTSWRR156 pKa = 11.84LCNSARR162 pKa = 11.84SNQSEE167 pKa = 4.31PPLDD171 pKa = 4.82RR172 pKa = 11.84IQEE175 pKa = 3.99VDD177 pKa = 4.03EE178 pKa = 5.68GDD180 pKa = 4.39DD181 pKa = 3.9NQGDD185 pKa = 4.32DD186 pKa = 4.06VNPAAAHH193 pKa = 5.37SPSRR197 pKa = 11.84NANHH201 pKa = 5.35QQQNGGRR208 pKa = 11.84TEE210 pKa = 4.26AEE212 pKa = 3.99LALEE216 pKa = 4.15QALEE220 pKa = 3.96ASNVEE225 pKa = 4.24EE226 pKa = 3.76QRR228 pKa = 11.84RR229 pKa = 11.84AYY231 pKa = 10.77VEE233 pKa = 4.09GGVLHH238 pKa = 6.19STTTLAPGQRR248 pKa = 11.84PEE250 pKa = 4.43LVEE253 pKa = 4.02PPGLEE258 pKa = 4.22GGCASGTRR266 pKa = 11.84QARR269 pKa = 11.84ARR271 pKa = 11.84FPQITSQPNFLHH283 pKa = 7.0SNNEE287 pKa = 3.66QNLQMAHH294 pKa = 6.77DD295 pKa = 4.05MRR297 pKa = 11.84NVGVGDD303 pKa = 4.16HH304 pKa = 6.83NPLQSEE310 pKa = 4.47VQTRR314 pKa = 11.84DD315 pKa = 3.17RR316 pKa = 11.84LVEE319 pKa = 3.85LLKK322 pKa = 10.9QKK324 pKa = 10.38VFTPGRR330 pKa = 11.84LRR332 pKa = 11.84KK333 pKa = 10.14AMVGFEE339 pKa = 4.23SARR342 pKa = 11.84VSALPKK348 pKa = 10.18KK349 pKa = 10.43LSEE352 pKa = 4.22EE353 pKa = 3.42EE354 pKa = 3.91RR355 pKa = 11.84MRR357 pKa = 11.84VEE359 pKa = 4.24LEE361 pKa = 3.83AMNAALSDD369 pKa = 4.06DD370 pKa = 3.77GVGFDD375 pKa = 3.77TVIKK379 pKa = 10.49AFVKK383 pKa = 10.84SEE385 pKa = 3.74VTAKK389 pKa = 10.63NKK391 pKa = 8.89PRR393 pKa = 11.84PIANHH398 pKa = 5.46GNVRR402 pKa = 11.84LYY404 pKa = 11.29ALAKK408 pKa = 8.44VAYY411 pKa = 9.07AFEE414 pKa = 4.46HH415 pKa = 6.51VMFEE419 pKa = 4.3TFKK422 pKa = 11.07GGSIKK427 pKa = 10.68GRR429 pKa = 11.84GKK431 pKa = 10.39KK432 pKa = 8.81EE433 pKa = 3.96AIEE436 pKa = 3.96EE437 pKa = 4.3LFSNMSNMKK446 pKa = 9.73NGRR449 pKa = 11.84WVEE452 pKa = 3.72NDD454 pKa = 2.97LTSFEE459 pKa = 4.53FGISEE464 pKa = 4.16PLKK467 pKa = 10.27QIEE470 pKa = 3.93MDD472 pKa = 3.34IFYY475 pKa = 9.97HH476 pKa = 5.46IAKK479 pKa = 9.84IIGVADD485 pKa = 3.98TGALLFEE492 pKa = 4.65RR493 pKa = 11.84VSNDD497 pKa = 3.12RR498 pKa = 11.84DD499 pKa = 3.4KK500 pKa = 11.55CATWKK505 pKa = 10.68LIYY508 pKa = 10.1TDD510 pKa = 3.15EE511 pKa = 4.5TGQKK515 pKa = 8.38RR516 pKa = 11.84TAKK519 pKa = 10.08IKK521 pKa = 10.17IPQTMRR527 pKa = 11.84EE528 pKa = 3.95SGDD531 pKa = 3.37RR532 pKa = 11.84VTSSGNFLQNLIAWFCFLVDD552 pKa = 3.56PDD554 pKa = 4.08YY555 pKa = 11.88VEE557 pKa = 5.48DD558 pKa = 4.34ALEE561 pKa = 3.94SLIKK565 pKa = 10.6HH566 pKa = 6.21RR567 pKa = 11.84GSKK570 pKa = 10.18LFYY573 pKa = 9.38VSRR576 pKa = 11.84RR577 pKa = 11.84DD578 pKa = 3.09RR579 pKa = 11.84STIEE583 pKa = 3.82LRR585 pKa = 11.84GKK587 pKa = 7.62QVRR590 pKa = 11.84KK591 pKa = 10.19KK592 pKa = 10.75YY593 pKa = 9.56LACLAFEE600 pKa = 5.22GDD602 pKa = 3.83DD603 pKa = 3.25TSGRR607 pKa = 11.84IEE609 pKa = 4.05EE610 pKa = 4.41PVWLNGPYY618 pKa = 10.55NPDD621 pKa = 3.14NSEE624 pKa = 4.06TKK626 pKa = 10.46DD627 pKa = 3.46DD628 pKa = 4.27CLVSSFFRR636 pKa = 11.84RR637 pKa = 11.84WGWKK641 pKa = 9.75AKK643 pKa = 10.3LIWKK647 pKa = 8.09PQEE650 pKa = 3.21GDD652 pKa = 3.16AYY654 pKa = 10.87VRR656 pKa = 11.84FVGYY660 pKa = 9.12EE661 pKa = 3.87ALIHH665 pKa = 6.61DD666 pKa = 4.62SKK668 pKa = 10.95IVYY671 pKa = 10.12DD672 pKa = 3.85GAEE675 pKa = 3.66MVMTPEE681 pKa = 4.71ARR683 pKa = 11.84RR684 pKa = 11.84LLNTKK689 pKa = 8.93SWTTTAVTPAEE700 pKa = 4.52LKK702 pKa = 8.98TCIRR706 pKa = 11.84VYY708 pKa = 10.5AASLADD714 pKa = 3.31GFKK717 pKa = 10.42RR718 pKa = 11.84VEE720 pKa = 3.73PMYY723 pKa = 11.2AFLNAVFDD731 pKa = 4.44SNKK734 pKa = 10.52GGVDD738 pKa = 3.09VDD740 pKa = 3.62AQKK743 pKa = 11.11VKK745 pKa = 10.58EE746 pKa = 3.98YY747 pKa = 10.6LLAVEE752 pKa = 5.45GEE754 pKa = 4.32MPEE757 pKa = 3.73HH758 pKa = 6.63GRR760 pKa = 11.84TVNCPIDD767 pKa = 3.57MPPFEE772 pKa = 6.7GGDD775 pKa = 3.22ADD777 pKa = 3.05KK778 pKa = 10.65WKK780 pKa = 10.67RR781 pKa = 11.84LLRR784 pKa = 11.84RR785 pKa = 11.84SAGDD789 pKa = 3.12FDD791 pKa = 3.75EE792 pKa = 6.18RR793 pKa = 11.84EE794 pKa = 4.14WATMCHH800 pKa = 6.57ISTIDD805 pKa = 3.06IHH807 pKa = 8.6GEE809 pKa = 3.64DD810 pKa = 4.16LAISVPAEE818 pKa = 3.59WRR820 pKa = 11.84NN821 pKa = 3.21
Molecular weight: 92.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
821 |
821 |
821 |
821.0 |
92.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.161 ± 0.0 | 2.68 ± 0.0 |
6.09 ± 0.0 | 7.917 ± 0.0 |
4.507 ± 0.0 | 6.699 ± 0.0 |
2.314 ± 0.0 | 4.019 ± 0.0 |
5.603 ± 0.0 | 8.039 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.436 ± 0.0 | 4.507 ± 0.0 |
4.507 ± 0.0 | 3.654 ± 0.0 |
7.186 ± 0.0 | 6.334 ± 0.0 |
5.238 ± 0.0 | 6.334 ± 0.0 |
1.705 ± 0.0 | 2.071 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
