
Candidatus Bipolaricaulis anaerobius
Taxonomy: cellular organisms; Bacteria; Bacteria incertae sedis; Bacteria candidate phyla; Candidatus Bipolaricaulota; Candidatus Bipolaricaulis
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins
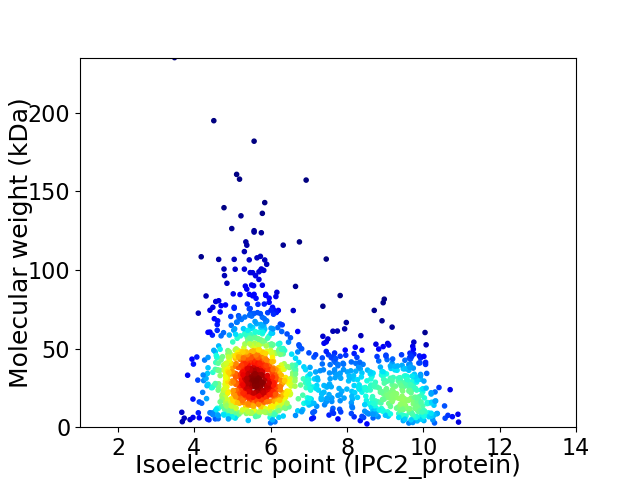
Virtual 2D-PAGE plot for 1319 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2X3L1W7|A0A2X3L1W7_9BACT 4-alpha-glucanotransferase OS=Candidatus Bipolaricaulis anaerobius OX=2026885 GN=malQ PE=3 SV=1
MM1 pKa = 7.33NWFSCGNRR9 pKa = 11.84DD10 pKa = 3.49ARR12 pKa = 11.84GHH14 pKa = 6.08AGHH17 pKa = 7.35RR18 pKa = 11.84SRR20 pKa = 11.84RR21 pKa = 11.84VLIPVLGLVFALGLIGFGATNTTTTLTSDD50 pKa = 3.74VNPSVHH56 pKa = 5.75GQPVTFTATVNPVPDD71 pKa = 4.17GGTVIFKK78 pKa = 11.0NEE80 pKa = 3.76TSVLGSVTVDD90 pKa = 2.89TGTGKK95 pKa = 10.75ANFSTNSLSVGSHH108 pKa = 6.85SITAEE113 pKa = 3.85YY114 pKa = 10.68SGTTDD119 pKa = 3.61YY120 pKa = 11.49NPSTSSPLDD129 pKa = 3.31QKK131 pKa = 11.07VDD133 pKa = 3.47KK134 pKa = 11.06ASTTVSVGSSTNPSVTGQSVTFTATVSAVSPGSGTPTGTVDD175 pKa = 5.09FYY177 pKa = 11.92ASGNLIAADD186 pKa = 4.26CPLSGGTATCSRR198 pKa = 11.84SFAHH202 pKa = 6.67IEE204 pKa = 4.07SPVAVTAHH212 pKa = 5.87YY213 pKa = 10.96SGDD216 pKa = 3.78TNFNPSDD223 pKa = 3.66NVLNPLTQTVDD234 pKa = 3.27KK235 pKa = 11.07ASTTVTFIPSGSSWVGDD252 pKa = 3.75SYY254 pKa = 11.21TVSGEE259 pKa = 4.17VTVTSPGSGVPTGTVTVRR277 pKa = 11.84DD278 pKa = 4.13DD279 pKa = 3.5TGEE282 pKa = 3.89TCAANLVEE290 pKa = 4.74GAWSCFLSSSGAAGTRR306 pKa = 11.84TLTAVYY312 pKa = 10.05EE313 pKa = 4.3GDD315 pKa = 3.33ANYY318 pKa = 10.72AGSSGSTTHH327 pKa = 6.22QVTTVATTLALTDD340 pKa = 3.9EE341 pKa = 5.19PDD343 pKa = 3.46PSVVGGAVTLTATATAEE360 pKa = 4.25TGGGSPTGTVSFSIGTTPLGSAEE383 pKa = 4.23LVSSGTNKK391 pKa = 10.12SEE393 pKa = 3.93AVLVTTQIPFGTHH406 pKa = 6.97TITATYY412 pKa = 9.61PGDD415 pKa = 3.34GRR417 pKa = 11.84FAGSTATAEE426 pKa = 4.12HH427 pKa = 6.26TASKK431 pKa = 10.87RR432 pKa = 11.84EE433 pKa = 3.98TTTLVMGSDD442 pKa = 3.5TALVVGDD449 pKa = 3.95TVTVTVRR456 pKa = 11.84VVDD459 pKa = 4.15TSPGASSMPTGTVSVSVSPTDD480 pKa = 3.28QGTPTSWSHH489 pKa = 6.44ALVAADD495 pKa = 3.41AGEE498 pKa = 4.19FTFTYY503 pKa = 9.72TPTSGEE509 pKa = 4.36TTPHH513 pKa = 6.53TFTATYY519 pKa = 10.39AGDD522 pKa = 3.87STHH525 pKa = 6.6SGSSGSFAQAIIKK538 pKa = 9.96RR539 pKa = 11.84AADD542 pKa = 3.26IQLVLDD548 pKa = 3.72PTTAYY553 pKa = 9.74ILQNVACQVHH563 pKa = 6.54VEE565 pKa = 4.35DD566 pKa = 4.81DD567 pKa = 3.82TAAGTPITPSGDD579 pKa = 3.21VTFDD583 pKa = 3.55DD584 pKa = 4.68GGKK587 pKa = 9.89NGAFTVSGTPGTTWPLDD604 pKa = 3.33GSGNLSVIYY613 pKa = 10.06TPGAFDD619 pKa = 5.11AGTTTITATYY629 pKa = 9.17NGSAVHH635 pKa = 6.39IGKK638 pKa = 8.26STTQLLTVLLRR649 pKa = 11.84PTEE652 pKa = 4.13VTVNGCTNTILVNQEE667 pKa = 3.26CSYY670 pKa = 10.51TVTVEE675 pKa = 4.09EE676 pKa = 4.64ADD678 pKa = 4.74GIPGTATAPFGTLSYY693 pKa = 10.89SSYY696 pKa = 10.49LGSDD700 pKa = 3.31ATQVPNSGAAPSGTFTYY717 pKa = 9.98TCLGLDD723 pKa = 3.61GHH725 pKa = 7.15AGIDD729 pKa = 3.57TIYY732 pKa = 11.37AHH734 pKa = 5.8YY735 pKa = 9.0TPNDD739 pKa = 4.63GIHH742 pKa = 6.21AASGGGFGQAIQRR755 pKa = 11.84RR756 pKa = 11.84PTITTVTGSGTPAGVTYY773 pKa = 9.08TATVQEE779 pKa = 4.5DD780 pKa = 3.68SGNAGTDD787 pKa = 3.32TILQGNIHH795 pKa = 7.08LLQPNEE801 pKa = 4.12YY802 pKa = 8.87PCTGLSGATLTCTGNFNTTSPLVNVTVQFGPTDD835 pKa = 3.69RR836 pKa = 11.84THH838 pKa = 6.87LGSTGSVNIEE848 pKa = 4.48RR849 pKa = 11.84YY850 pKa = 9.83DD851 pKa = 3.82QFPDD855 pKa = 3.69DD856 pKa = 5.37LGDD859 pKa = 3.88GSDD862 pKa = 4.92GSEE865 pKa = 4.36CDD867 pKa = 4.14DD868 pKa = 4.02GCGSGGKK875 pKa = 9.21NVAQMIFDD883 pKa = 4.72LNASEE888 pKa = 4.76VALQAVKK895 pKa = 10.81LGLDD899 pKa = 3.35TTALVLDD906 pKa = 4.77VIPDD910 pKa = 3.66GVITGGVVVQTGVTIPYY927 pKa = 10.03SDD929 pKa = 3.17IAKK932 pKa = 10.24AVVAGAGILLDD943 pKa = 3.61IAIMSMDD950 pKa = 4.15TDD952 pKa = 4.66LDD954 pKa = 4.38DD955 pKa = 6.82DD956 pKa = 4.49GLPDD960 pKa = 3.61VLEE963 pKa = 4.47EE964 pKa = 4.21NVTDD968 pKa = 3.55TDD970 pKa = 3.71YY971 pKa = 11.81DD972 pKa = 3.28KK973 pKa = 11.13WDD975 pKa = 3.61TDD977 pKa = 3.73GDD979 pKa = 4.32GMGDD983 pKa = 3.13WDD985 pKa = 4.33EE986 pKa = 5.13LEE988 pKa = 4.38EE989 pKa = 4.65ASGYY993 pKa = 10.29YY994 pKa = 10.17GGSRR998 pKa = 11.84RR999 pKa = 11.84PDD1001 pKa = 3.76PNDD1004 pKa = 3.39SDD1006 pKa = 5.17SDD1008 pKa = 4.09DD1009 pKa = 5.32DD1010 pKa = 5.27GIPDD1014 pKa = 4.3GDD1016 pKa = 4.06EE1017 pKa = 3.7QSLTHH1022 pKa = 6.43TNFCVADD1029 pKa = 4.0TDD1031 pKa = 4.2CDD1033 pKa = 3.93TVIDD1037 pKa = 4.53GGQGLAAVGEE1047 pKa = 4.27AEE1049 pKa = 4.02TWGFADD1055 pKa = 4.68IRR1057 pKa = 11.84DD1058 pKa = 3.91HH1059 pKa = 7.43ADD1061 pKa = 3.12PLMEE1065 pKa = 4.4EE1066 pKa = 4.2TDD1068 pKa = 3.87GDD1070 pKa = 4.12GLRR1073 pKa = 11.84DD1074 pKa = 3.88DD1075 pKa = 4.65VEE1077 pKa = 4.3IAAGCPFVNDD1087 pKa = 5.04DD1088 pKa = 5.34DD1089 pKa = 6.23SDD1091 pKa = 5.11DD1092 pKa = 5.94DD1093 pKa = 4.14GLQDD1097 pKa = 5.67GFEE1100 pKa = 4.29SWDD1103 pKa = 3.59GNGTCVTGDD1112 pKa = 3.1IGDD1115 pKa = 4.61SLTQSSLTGEE1125 pKa = 4.31TDD1127 pKa = 3.45FCDD1130 pKa = 4.4PDD1132 pKa = 3.61TDD1134 pKa = 4.88GDD1136 pKa = 4.12GLTDD1140 pKa = 3.82GEE1142 pKa = 4.52EE1143 pKa = 4.1VALLGGLPVTSSGFKK1158 pKa = 9.49PVAPRR1163 pKa = 11.84GVSTVFGQDD1172 pKa = 2.97GSALTATIPALDD1184 pKa = 5.15DD1185 pKa = 5.57DD1186 pKa = 4.98SDD1188 pKa = 4.29NDD1190 pKa = 3.94GLSDD1194 pKa = 4.0CEE1196 pKa = 4.2EE1197 pKa = 4.08VSITHH1202 pKa = 6.55TNPLDD1207 pKa = 3.52QDD1209 pKa = 3.69SDD1211 pKa = 3.75NDD1213 pKa = 4.16TLSDD1217 pKa = 3.96ANEE1220 pKa = 4.54LIAVVGSSWPNRR1232 pKa = 11.84SFIQVSDD1239 pKa = 4.11PLDD1242 pKa = 4.38PDD1244 pKa = 3.84SDD1246 pKa = 4.75DD1247 pKa = 5.96DD1248 pKa = 4.16GLPDD1252 pKa = 3.38QVEE1255 pKa = 4.46YY1256 pKa = 10.24PGSGLGMLRR1265 pKa = 11.84TSGGVPDD1272 pKa = 4.36NVCSYY1277 pKa = 11.37VGDD1280 pKa = 4.92DD1281 pKa = 5.43DD1282 pKa = 6.56SDD1284 pKa = 5.06DD1285 pKa = 5.64DD1286 pKa = 4.31GLQDD1290 pKa = 4.07GHH1292 pKa = 6.88EE1293 pKa = 4.92DD1294 pKa = 3.85LDD1296 pKa = 4.58KK1297 pKa = 11.44DD1298 pKa = 3.95GQFDD1302 pKa = 4.39LGTIGTTQTIGTGEE1316 pKa = 4.26TNLCNPDD1323 pKa = 3.34TDD1325 pKa = 5.33GDD1327 pKa = 4.16GLDD1330 pKa = 4.72DD1331 pKa = 5.57GQEE1334 pKa = 3.87EE1335 pKa = 4.47WQLGAGSVAVTSQDD1349 pKa = 3.16SATIAHH1355 pKa = 6.42TVPALDD1361 pKa = 4.54MDD1363 pKa = 4.66SDD1365 pKa = 5.04DD1366 pKa = 5.89DD1367 pKa = 4.14GLSDD1371 pKa = 3.87GEE1373 pKa = 4.51EE1374 pKa = 4.19VNVTHH1379 pKa = 7.23TDD1381 pKa = 3.5PLDD1384 pKa = 3.38WDD1386 pKa = 4.03TDD1388 pKa = 3.46NDD1390 pKa = 4.26TLSDD1394 pKa = 4.09LNEE1397 pKa = 4.09LLVTGGTWPQRR1408 pKa = 11.84TFSQVSDD1415 pKa = 4.19PLDD1418 pKa = 4.63PDD1420 pKa = 3.61TDD1422 pKa = 4.28DD1423 pKa = 5.79DD1424 pKa = 4.37GLIDD1428 pKa = 3.74SVEE1431 pKa = 4.07YY1432 pKa = 11.02NGTGSDD1438 pKa = 3.55RR1439 pKa = 11.84FLHH1442 pKa = 6.62LSTDD1446 pKa = 3.56GVDD1449 pKa = 5.49DD1450 pKa = 4.34LVCPYY1455 pKa = 11.14VNDD1458 pKa = 5.39DD1459 pKa = 5.01DD1460 pKa = 6.51SDD1462 pKa = 4.98DD1463 pKa = 6.16DD1464 pKa = 4.13GLQDD1468 pKa = 3.69GGEE1471 pKa = 4.28DD1472 pKa = 3.71ANHH1475 pKa = 7.03DD1476 pKa = 3.62GMLGVGGSGISIGDD1490 pKa = 3.47WGSQATKK1497 pKa = 10.63SVDD1500 pKa = 3.11YY1501 pKa = 10.37WEE1503 pKa = 5.8CNLCNPDD1510 pKa = 3.32TDD1512 pKa = 4.77GDD1514 pKa = 4.09GLLDD1518 pKa = 3.96GEE1520 pKa = 4.69EE1521 pKa = 4.17VALLGGGPILGRR1533 pKa = 11.84PRR1535 pKa = 11.84PIPGFNTVIPEE1546 pKa = 4.12AVSTVLPLGTGPAGSPPGTRR1566 pKa = 11.84NSQLGPYY1573 pKa = 8.7TFVPAAGASMIATIPALDD1591 pKa = 4.09NDD1593 pKa = 4.24SDD1595 pKa = 4.36NDD1597 pKa = 3.87GLSDD1601 pKa = 3.85YY1602 pKa = 11.2EE1603 pKa = 4.25EE1604 pKa = 4.4VNITGTDD1611 pKa = 3.7PLDD1614 pKa = 3.52QDD1616 pKa = 4.02SDD1618 pKa = 3.77NDD1620 pKa = 3.81TLMDD1624 pKa = 4.3SDD1626 pKa = 4.47EE1627 pKa = 5.3LIAIAGTWPQRR1638 pKa = 11.84TFDD1641 pKa = 3.89QEE1643 pKa = 4.84SNPLDD1648 pKa = 3.7INTDD1652 pKa = 3.63DD1653 pKa = 4.86DD1654 pKa = 4.67HH1655 pKa = 8.76LFDD1658 pKa = 4.37PQEE1661 pKa = 3.99FSGSGLSTLAGALGGTRR1678 pKa = 11.84DD1679 pKa = 3.75MQCPFVNDD1687 pKa = 4.69DD1688 pKa = 4.71DD1689 pKa = 6.24SDD1691 pKa = 4.82DD1692 pKa = 6.07DD1693 pKa = 5.15GIQDD1697 pKa = 3.43GAVVVRR1703 pKa = 11.84TFVAVGVTYY1712 pKa = 10.65SWTHH1716 pKa = 5.95YY1717 pKa = 10.94EE1718 pKa = 4.28DD1719 pKa = 4.15FADD1722 pKa = 3.69VTGATDD1728 pKa = 3.97AAPGTVRR1735 pKa = 11.84TVVTPAGGEE1744 pKa = 4.09QNDD1747 pKa = 3.53DD1748 pKa = 4.52SIWNVCDD1755 pKa = 4.13PDD1757 pKa = 4.13SDD1759 pKa = 4.65GDD1761 pKa = 3.94GLLDD1765 pKa = 3.9GEE1767 pKa = 4.62EE1768 pKa = 4.28VAIGTDD1774 pKa = 3.28PGDD1777 pKa = 3.48WDD1779 pKa = 3.88TDD1781 pKa = 3.64DD1782 pKa = 5.72DD1783 pKa = 5.45GRR1785 pKa = 11.84NDD1787 pKa = 2.77WHH1789 pKa = 6.45EE1790 pKa = 4.02QTGGGPIPTDD1800 pKa = 3.6PFDD1803 pKa = 5.9PDD1805 pKa = 3.46TDD1807 pKa = 4.32DD1808 pKa = 6.71DD1809 pKa = 4.54GLLDD1813 pKa = 3.62SAEE1816 pKa = 4.27VFGSNPTNPANCDD1829 pKa = 2.79TDD1831 pKa = 5.29ADD1833 pKa = 4.28GLCDD1837 pKa = 3.6GGARR1841 pKa = 11.84TPYY1844 pKa = 7.58MTSGHH1849 pKa = 6.53PSVVVSPRR1857 pKa = 11.84CRR1859 pKa = 11.84TGIGGHH1865 pKa = 6.4PNPLGIGEE1873 pKa = 5.11DD1874 pKa = 3.89EE1875 pKa = 5.32DD1876 pKa = 6.36GDD1878 pKa = 4.42GAWDD1882 pKa = 4.35SGEE1885 pKa = 4.09TDD1887 pKa = 4.11PNNPDD1892 pKa = 3.17TDD1894 pKa = 3.64GDD1896 pKa = 4.09AVGDD1900 pKa = 3.78GFEE1903 pKa = 4.15RR1904 pKa = 11.84LSFSVSRR1911 pKa = 11.84QSLIPATDD1919 pKa = 3.57LFGRR1923 pKa = 11.84PITVVYY1929 pKa = 10.29PEE1931 pKa = 4.48ANNVKK1936 pKa = 9.29PACGCMNPLTSDD1948 pKa = 3.27SDD1950 pKa = 3.76EE1951 pKa = 5.37DD1952 pKa = 3.82GLLDD1956 pKa = 4.53GEE1958 pKa = 4.68EE1959 pKa = 4.45DD1960 pKa = 3.98LNHH1963 pKa = 7.23DD1964 pKa = 3.75GHH1966 pKa = 7.56FDD1968 pKa = 3.95FLPSDD1973 pKa = 4.29FDD1975 pKa = 4.29YY1976 pKa = 10.95GHH1978 pKa = 7.32HH1979 pKa = 6.47GPIPGPAYY1987 pKa = 10.19GSPLEE1992 pKa = 4.56TNPCDD1997 pKa = 4.41PDD1999 pKa = 3.69TDD2001 pKa = 4.28HH2002 pKa = 7.99DD2003 pKa = 4.35GLTDD2007 pKa = 3.79LEE2009 pKa = 4.37EE2010 pKa = 5.08LRR2012 pKa = 11.84GQANPPSFFPFNPTDD2027 pKa = 4.2PLDD2030 pKa = 4.34HH2031 pKa = 7.29DD2032 pKa = 4.7TDD2034 pKa = 4.01NDD2036 pKa = 4.2WIYY2039 pKa = 11.02DD2040 pKa = 3.75GPEE2043 pKa = 3.83VRR2045 pKa = 11.84WVCTEE2050 pKa = 3.95LLFVSLDD2057 pKa = 3.41NDD2059 pKa = 3.44GDD2061 pKa = 4.05TFIDD2065 pKa = 3.96EE2066 pKa = 4.77DD2067 pKa = 4.36PVDD2070 pKa = 4.1EE2071 pKa = 4.83VDD2073 pKa = 3.94NDD2075 pKa = 3.74GDD2077 pKa = 4.14GLIDD2081 pKa = 3.95EE2082 pKa = 5.76DD2083 pKa = 5.59DD2084 pKa = 3.34VDD2086 pKa = 4.59FYY2088 pKa = 12.1VRR2090 pKa = 11.84FVPVLDD2096 pKa = 3.58PTNRR2100 pKa = 11.84DD2101 pKa = 3.02SDD2103 pKa = 3.81SDD2105 pKa = 4.06GFIDD2109 pKa = 6.2GLDD2112 pKa = 3.63EE2113 pKa = 5.4DD2114 pKa = 4.64PCNTQLIPIARR2125 pKa = 11.84PVVQTPVDD2133 pKa = 3.45TDD2135 pKa = 3.49GDD2137 pKa = 4.02GFADD2141 pKa = 4.45DD2142 pKa = 5.62DD2143 pKa = 4.61EE2144 pKa = 5.42IAAGTHH2150 pKa = 6.42PNDD2153 pKa = 3.72PTSFPCAFGSDD2164 pKa = 3.72LDD2166 pKa = 4.44LDD2168 pKa = 4.62GVCDD2172 pKa = 4.08DD2173 pKa = 5.15RR2174 pKa = 11.84LWLTDD2179 pKa = 3.48TNHH2182 pKa = 7.34DD2183 pKa = 3.71GTADD2187 pKa = 3.47TVTIDD2192 pKa = 2.76IDD2194 pKa = 4.1TNVLVDD2200 pKa = 4.15LRR2202 pKa = 11.84VEE2204 pKa = 3.84ILSRR2208 pKa = 11.84DD2209 pKa = 3.7LQRR2212 pKa = 11.84GDD2214 pKa = 3.73FDD2216 pKa = 6.02GDD2218 pKa = 3.53GQADD2222 pKa = 3.82DD2223 pKa = 3.65GRR2225 pKa = 11.84YY2226 pKa = 7.14TISYY2230 pKa = 9.16AFANRR2235 pKa = 11.84RR2236 pKa = 11.84VIQPRR2241 pKa = 11.84LTVVILDD2248 pKa = 3.77YY2249 pKa = 11.59GCDD2252 pKa = 3.47LRR2254 pKa = 11.84VDD2256 pKa = 3.66QVEE2259 pKa = 4.36VTKK2262 pKa = 11.08
MM1 pKa = 7.33NWFSCGNRR9 pKa = 11.84DD10 pKa = 3.49ARR12 pKa = 11.84GHH14 pKa = 6.08AGHH17 pKa = 7.35RR18 pKa = 11.84SRR20 pKa = 11.84RR21 pKa = 11.84VLIPVLGLVFALGLIGFGATNTTTTLTSDD50 pKa = 3.74VNPSVHH56 pKa = 5.75GQPVTFTATVNPVPDD71 pKa = 4.17GGTVIFKK78 pKa = 11.0NEE80 pKa = 3.76TSVLGSVTVDD90 pKa = 2.89TGTGKK95 pKa = 10.75ANFSTNSLSVGSHH108 pKa = 6.85SITAEE113 pKa = 3.85YY114 pKa = 10.68SGTTDD119 pKa = 3.61YY120 pKa = 11.49NPSTSSPLDD129 pKa = 3.31QKK131 pKa = 11.07VDD133 pKa = 3.47KK134 pKa = 11.06ASTTVSVGSSTNPSVTGQSVTFTATVSAVSPGSGTPTGTVDD175 pKa = 5.09FYY177 pKa = 11.92ASGNLIAADD186 pKa = 4.26CPLSGGTATCSRR198 pKa = 11.84SFAHH202 pKa = 6.67IEE204 pKa = 4.07SPVAVTAHH212 pKa = 5.87YY213 pKa = 10.96SGDD216 pKa = 3.78TNFNPSDD223 pKa = 3.66NVLNPLTQTVDD234 pKa = 3.27KK235 pKa = 11.07ASTTVTFIPSGSSWVGDD252 pKa = 3.75SYY254 pKa = 11.21TVSGEE259 pKa = 4.17VTVTSPGSGVPTGTVTVRR277 pKa = 11.84DD278 pKa = 4.13DD279 pKa = 3.5TGEE282 pKa = 3.89TCAANLVEE290 pKa = 4.74GAWSCFLSSSGAAGTRR306 pKa = 11.84TLTAVYY312 pKa = 10.05EE313 pKa = 4.3GDD315 pKa = 3.33ANYY318 pKa = 10.72AGSSGSTTHH327 pKa = 6.22QVTTVATTLALTDD340 pKa = 3.9EE341 pKa = 5.19PDD343 pKa = 3.46PSVVGGAVTLTATATAEE360 pKa = 4.25TGGGSPTGTVSFSIGTTPLGSAEE383 pKa = 4.23LVSSGTNKK391 pKa = 10.12SEE393 pKa = 3.93AVLVTTQIPFGTHH406 pKa = 6.97TITATYY412 pKa = 9.61PGDD415 pKa = 3.34GRR417 pKa = 11.84FAGSTATAEE426 pKa = 4.12HH427 pKa = 6.26TASKK431 pKa = 10.87RR432 pKa = 11.84EE433 pKa = 3.98TTTLVMGSDD442 pKa = 3.5TALVVGDD449 pKa = 3.95TVTVTVRR456 pKa = 11.84VVDD459 pKa = 4.15TSPGASSMPTGTVSVSVSPTDD480 pKa = 3.28QGTPTSWSHH489 pKa = 6.44ALVAADD495 pKa = 3.41AGEE498 pKa = 4.19FTFTYY503 pKa = 9.72TPTSGEE509 pKa = 4.36TTPHH513 pKa = 6.53TFTATYY519 pKa = 10.39AGDD522 pKa = 3.87STHH525 pKa = 6.6SGSSGSFAQAIIKK538 pKa = 9.96RR539 pKa = 11.84AADD542 pKa = 3.26IQLVLDD548 pKa = 3.72PTTAYY553 pKa = 9.74ILQNVACQVHH563 pKa = 6.54VEE565 pKa = 4.35DD566 pKa = 4.81DD567 pKa = 3.82TAAGTPITPSGDD579 pKa = 3.21VTFDD583 pKa = 3.55DD584 pKa = 4.68GGKK587 pKa = 9.89NGAFTVSGTPGTTWPLDD604 pKa = 3.33GSGNLSVIYY613 pKa = 10.06TPGAFDD619 pKa = 5.11AGTTTITATYY629 pKa = 9.17NGSAVHH635 pKa = 6.39IGKK638 pKa = 8.26STTQLLTVLLRR649 pKa = 11.84PTEE652 pKa = 4.13VTVNGCTNTILVNQEE667 pKa = 3.26CSYY670 pKa = 10.51TVTVEE675 pKa = 4.09EE676 pKa = 4.64ADD678 pKa = 4.74GIPGTATAPFGTLSYY693 pKa = 10.89SSYY696 pKa = 10.49LGSDD700 pKa = 3.31ATQVPNSGAAPSGTFTYY717 pKa = 9.98TCLGLDD723 pKa = 3.61GHH725 pKa = 7.15AGIDD729 pKa = 3.57TIYY732 pKa = 11.37AHH734 pKa = 5.8YY735 pKa = 9.0TPNDD739 pKa = 4.63GIHH742 pKa = 6.21AASGGGFGQAIQRR755 pKa = 11.84RR756 pKa = 11.84PTITTVTGSGTPAGVTYY773 pKa = 9.08TATVQEE779 pKa = 4.5DD780 pKa = 3.68SGNAGTDD787 pKa = 3.32TILQGNIHH795 pKa = 7.08LLQPNEE801 pKa = 4.12YY802 pKa = 8.87PCTGLSGATLTCTGNFNTTSPLVNVTVQFGPTDD835 pKa = 3.69RR836 pKa = 11.84THH838 pKa = 6.87LGSTGSVNIEE848 pKa = 4.48RR849 pKa = 11.84YY850 pKa = 9.83DD851 pKa = 3.82QFPDD855 pKa = 3.69DD856 pKa = 5.37LGDD859 pKa = 3.88GSDD862 pKa = 4.92GSEE865 pKa = 4.36CDD867 pKa = 4.14DD868 pKa = 4.02GCGSGGKK875 pKa = 9.21NVAQMIFDD883 pKa = 4.72LNASEE888 pKa = 4.76VALQAVKK895 pKa = 10.81LGLDD899 pKa = 3.35TTALVLDD906 pKa = 4.77VIPDD910 pKa = 3.66GVITGGVVVQTGVTIPYY927 pKa = 10.03SDD929 pKa = 3.17IAKK932 pKa = 10.24AVVAGAGILLDD943 pKa = 3.61IAIMSMDD950 pKa = 4.15TDD952 pKa = 4.66LDD954 pKa = 4.38DD955 pKa = 6.82DD956 pKa = 4.49GLPDD960 pKa = 3.61VLEE963 pKa = 4.47EE964 pKa = 4.21NVTDD968 pKa = 3.55TDD970 pKa = 3.71YY971 pKa = 11.81DD972 pKa = 3.28KK973 pKa = 11.13WDD975 pKa = 3.61TDD977 pKa = 3.73GDD979 pKa = 4.32GMGDD983 pKa = 3.13WDD985 pKa = 4.33EE986 pKa = 5.13LEE988 pKa = 4.38EE989 pKa = 4.65ASGYY993 pKa = 10.29YY994 pKa = 10.17GGSRR998 pKa = 11.84RR999 pKa = 11.84PDD1001 pKa = 3.76PNDD1004 pKa = 3.39SDD1006 pKa = 5.17SDD1008 pKa = 4.09DD1009 pKa = 5.32DD1010 pKa = 5.27GIPDD1014 pKa = 4.3GDD1016 pKa = 4.06EE1017 pKa = 3.7QSLTHH1022 pKa = 6.43TNFCVADD1029 pKa = 4.0TDD1031 pKa = 4.2CDD1033 pKa = 3.93TVIDD1037 pKa = 4.53GGQGLAAVGEE1047 pKa = 4.27AEE1049 pKa = 4.02TWGFADD1055 pKa = 4.68IRR1057 pKa = 11.84DD1058 pKa = 3.91HH1059 pKa = 7.43ADD1061 pKa = 3.12PLMEE1065 pKa = 4.4EE1066 pKa = 4.2TDD1068 pKa = 3.87GDD1070 pKa = 4.12GLRR1073 pKa = 11.84DD1074 pKa = 3.88DD1075 pKa = 4.65VEE1077 pKa = 4.3IAAGCPFVNDD1087 pKa = 5.04DD1088 pKa = 5.34DD1089 pKa = 6.23SDD1091 pKa = 5.11DD1092 pKa = 5.94DD1093 pKa = 4.14GLQDD1097 pKa = 5.67GFEE1100 pKa = 4.29SWDD1103 pKa = 3.59GNGTCVTGDD1112 pKa = 3.1IGDD1115 pKa = 4.61SLTQSSLTGEE1125 pKa = 4.31TDD1127 pKa = 3.45FCDD1130 pKa = 4.4PDD1132 pKa = 3.61TDD1134 pKa = 4.88GDD1136 pKa = 4.12GLTDD1140 pKa = 3.82GEE1142 pKa = 4.52EE1143 pKa = 4.1VALLGGLPVTSSGFKK1158 pKa = 9.49PVAPRR1163 pKa = 11.84GVSTVFGQDD1172 pKa = 2.97GSALTATIPALDD1184 pKa = 5.15DD1185 pKa = 5.57DD1186 pKa = 4.98SDD1188 pKa = 4.29NDD1190 pKa = 3.94GLSDD1194 pKa = 4.0CEE1196 pKa = 4.2EE1197 pKa = 4.08VSITHH1202 pKa = 6.55TNPLDD1207 pKa = 3.52QDD1209 pKa = 3.69SDD1211 pKa = 3.75NDD1213 pKa = 4.16TLSDD1217 pKa = 3.96ANEE1220 pKa = 4.54LIAVVGSSWPNRR1232 pKa = 11.84SFIQVSDD1239 pKa = 4.11PLDD1242 pKa = 4.38PDD1244 pKa = 3.84SDD1246 pKa = 4.75DD1247 pKa = 5.96DD1248 pKa = 4.16GLPDD1252 pKa = 3.38QVEE1255 pKa = 4.46YY1256 pKa = 10.24PGSGLGMLRR1265 pKa = 11.84TSGGVPDD1272 pKa = 4.36NVCSYY1277 pKa = 11.37VGDD1280 pKa = 4.92DD1281 pKa = 5.43DD1282 pKa = 6.56SDD1284 pKa = 5.06DD1285 pKa = 5.64DD1286 pKa = 4.31GLQDD1290 pKa = 4.07GHH1292 pKa = 6.88EE1293 pKa = 4.92DD1294 pKa = 3.85LDD1296 pKa = 4.58KK1297 pKa = 11.44DD1298 pKa = 3.95GQFDD1302 pKa = 4.39LGTIGTTQTIGTGEE1316 pKa = 4.26TNLCNPDD1323 pKa = 3.34TDD1325 pKa = 5.33GDD1327 pKa = 4.16GLDD1330 pKa = 4.72DD1331 pKa = 5.57GQEE1334 pKa = 3.87EE1335 pKa = 4.47WQLGAGSVAVTSQDD1349 pKa = 3.16SATIAHH1355 pKa = 6.42TVPALDD1361 pKa = 4.54MDD1363 pKa = 4.66SDD1365 pKa = 5.04DD1366 pKa = 5.89DD1367 pKa = 4.14GLSDD1371 pKa = 3.87GEE1373 pKa = 4.51EE1374 pKa = 4.19VNVTHH1379 pKa = 7.23TDD1381 pKa = 3.5PLDD1384 pKa = 3.38WDD1386 pKa = 4.03TDD1388 pKa = 3.46NDD1390 pKa = 4.26TLSDD1394 pKa = 4.09LNEE1397 pKa = 4.09LLVTGGTWPQRR1408 pKa = 11.84TFSQVSDD1415 pKa = 4.19PLDD1418 pKa = 4.63PDD1420 pKa = 3.61TDD1422 pKa = 4.28DD1423 pKa = 5.79DD1424 pKa = 4.37GLIDD1428 pKa = 3.74SVEE1431 pKa = 4.07YY1432 pKa = 11.02NGTGSDD1438 pKa = 3.55RR1439 pKa = 11.84FLHH1442 pKa = 6.62LSTDD1446 pKa = 3.56GVDD1449 pKa = 5.49DD1450 pKa = 4.34LVCPYY1455 pKa = 11.14VNDD1458 pKa = 5.39DD1459 pKa = 5.01DD1460 pKa = 6.51SDD1462 pKa = 4.98DD1463 pKa = 6.16DD1464 pKa = 4.13GLQDD1468 pKa = 3.69GGEE1471 pKa = 4.28DD1472 pKa = 3.71ANHH1475 pKa = 7.03DD1476 pKa = 3.62GMLGVGGSGISIGDD1490 pKa = 3.47WGSQATKK1497 pKa = 10.63SVDD1500 pKa = 3.11YY1501 pKa = 10.37WEE1503 pKa = 5.8CNLCNPDD1510 pKa = 3.32TDD1512 pKa = 4.77GDD1514 pKa = 4.09GLLDD1518 pKa = 3.96GEE1520 pKa = 4.69EE1521 pKa = 4.17VALLGGGPILGRR1533 pKa = 11.84PRR1535 pKa = 11.84PIPGFNTVIPEE1546 pKa = 4.12AVSTVLPLGTGPAGSPPGTRR1566 pKa = 11.84NSQLGPYY1573 pKa = 8.7TFVPAAGASMIATIPALDD1591 pKa = 4.09NDD1593 pKa = 4.24SDD1595 pKa = 4.36NDD1597 pKa = 3.87GLSDD1601 pKa = 3.85YY1602 pKa = 11.2EE1603 pKa = 4.25EE1604 pKa = 4.4VNITGTDD1611 pKa = 3.7PLDD1614 pKa = 3.52QDD1616 pKa = 4.02SDD1618 pKa = 3.77NDD1620 pKa = 3.81TLMDD1624 pKa = 4.3SDD1626 pKa = 4.47EE1627 pKa = 5.3LIAIAGTWPQRR1638 pKa = 11.84TFDD1641 pKa = 3.89QEE1643 pKa = 4.84SNPLDD1648 pKa = 3.7INTDD1652 pKa = 3.63DD1653 pKa = 4.86DD1654 pKa = 4.67HH1655 pKa = 8.76LFDD1658 pKa = 4.37PQEE1661 pKa = 3.99FSGSGLSTLAGALGGTRR1678 pKa = 11.84DD1679 pKa = 3.75MQCPFVNDD1687 pKa = 4.69DD1688 pKa = 4.71DD1689 pKa = 6.24SDD1691 pKa = 4.82DD1692 pKa = 6.07DD1693 pKa = 5.15GIQDD1697 pKa = 3.43GAVVVRR1703 pKa = 11.84TFVAVGVTYY1712 pKa = 10.65SWTHH1716 pKa = 5.95YY1717 pKa = 10.94EE1718 pKa = 4.28DD1719 pKa = 4.15FADD1722 pKa = 3.69VTGATDD1728 pKa = 3.97AAPGTVRR1735 pKa = 11.84TVVTPAGGEE1744 pKa = 4.09QNDD1747 pKa = 3.53DD1748 pKa = 4.52SIWNVCDD1755 pKa = 4.13PDD1757 pKa = 4.13SDD1759 pKa = 4.65GDD1761 pKa = 3.94GLLDD1765 pKa = 3.9GEE1767 pKa = 4.62EE1768 pKa = 4.28VAIGTDD1774 pKa = 3.28PGDD1777 pKa = 3.48WDD1779 pKa = 3.88TDD1781 pKa = 3.64DD1782 pKa = 5.72DD1783 pKa = 5.45GRR1785 pKa = 11.84NDD1787 pKa = 2.77WHH1789 pKa = 6.45EE1790 pKa = 4.02QTGGGPIPTDD1800 pKa = 3.6PFDD1803 pKa = 5.9PDD1805 pKa = 3.46TDD1807 pKa = 4.32DD1808 pKa = 6.71DD1809 pKa = 4.54GLLDD1813 pKa = 3.62SAEE1816 pKa = 4.27VFGSNPTNPANCDD1829 pKa = 2.79TDD1831 pKa = 5.29ADD1833 pKa = 4.28GLCDD1837 pKa = 3.6GGARR1841 pKa = 11.84TPYY1844 pKa = 7.58MTSGHH1849 pKa = 6.53PSVVVSPRR1857 pKa = 11.84CRR1859 pKa = 11.84TGIGGHH1865 pKa = 6.4PNPLGIGEE1873 pKa = 5.11DD1874 pKa = 3.89EE1875 pKa = 5.32DD1876 pKa = 6.36GDD1878 pKa = 4.42GAWDD1882 pKa = 4.35SGEE1885 pKa = 4.09TDD1887 pKa = 4.11PNNPDD1892 pKa = 3.17TDD1894 pKa = 3.64GDD1896 pKa = 4.09AVGDD1900 pKa = 3.78GFEE1903 pKa = 4.15RR1904 pKa = 11.84LSFSVSRR1911 pKa = 11.84QSLIPATDD1919 pKa = 3.57LFGRR1923 pKa = 11.84PITVVYY1929 pKa = 10.29PEE1931 pKa = 4.48ANNVKK1936 pKa = 9.29PACGCMNPLTSDD1948 pKa = 3.27SDD1950 pKa = 3.76EE1951 pKa = 5.37DD1952 pKa = 3.82GLLDD1956 pKa = 4.53GEE1958 pKa = 4.68EE1959 pKa = 4.45DD1960 pKa = 3.98LNHH1963 pKa = 7.23DD1964 pKa = 3.75GHH1966 pKa = 7.56FDD1968 pKa = 3.95FLPSDD1973 pKa = 4.29FDD1975 pKa = 4.29YY1976 pKa = 10.95GHH1978 pKa = 7.32HH1979 pKa = 6.47GPIPGPAYY1987 pKa = 10.19GSPLEE1992 pKa = 4.56TNPCDD1997 pKa = 4.41PDD1999 pKa = 3.69TDD2001 pKa = 4.28HH2002 pKa = 7.99DD2003 pKa = 4.35GLTDD2007 pKa = 3.79LEE2009 pKa = 4.37EE2010 pKa = 5.08LRR2012 pKa = 11.84GQANPPSFFPFNPTDD2027 pKa = 4.2PLDD2030 pKa = 4.34HH2031 pKa = 7.29DD2032 pKa = 4.7TDD2034 pKa = 4.01NDD2036 pKa = 4.2WIYY2039 pKa = 11.02DD2040 pKa = 3.75GPEE2043 pKa = 3.83VRR2045 pKa = 11.84WVCTEE2050 pKa = 3.95LLFVSLDD2057 pKa = 3.41NDD2059 pKa = 3.44GDD2061 pKa = 4.05TFIDD2065 pKa = 3.96EE2066 pKa = 4.77DD2067 pKa = 4.36PVDD2070 pKa = 4.1EE2071 pKa = 4.83VDD2073 pKa = 3.94NDD2075 pKa = 3.74GDD2077 pKa = 4.14GLIDD2081 pKa = 3.95EE2082 pKa = 5.76DD2083 pKa = 5.59DD2084 pKa = 3.34VDD2086 pKa = 4.59FYY2088 pKa = 12.1VRR2090 pKa = 11.84FVPVLDD2096 pKa = 3.58PTNRR2100 pKa = 11.84DD2101 pKa = 3.02SDD2103 pKa = 3.81SDD2105 pKa = 4.06GFIDD2109 pKa = 6.2GLDD2112 pKa = 3.63EE2113 pKa = 5.4DD2114 pKa = 4.64PCNTQLIPIARR2125 pKa = 11.84PVVQTPVDD2133 pKa = 3.45TDD2135 pKa = 3.49GDD2137 pKa = 4.02GFADD2141 pKa = 4.45DD2142 pKa = 5.62DD2143 pKa = 4.61EE2144 pKa = 5.42IAAGTHH2150 pKa = 6.42PNDD2153 pKa = 3.72PTSFPCAFGSDD2164 pKa = 3.72LDD2166 pKa = 4.44LDD2168 pKa = 4.62GVCDD2172 pKa = 4.08DD2173 pKa = 5.15RR2174 pKa = 11.84LWLTDD2179 pKa = 3.48TNHH2182 pKa = 7.34DD2183 pKa = 3.71GTADD2187 pKa = 3.47TVTIDD2192 pKa = 2.76IDD2194 pKa = 4.1TNVLVDD2200 pKa = 4.15LRR2202 pKa = 11.84VEE2204 pKa = 3.84ILSRR2208 pKa = 11.84DD2209 pKa = 3.7LQRR2212 pKa = 11.84GDD2214 pKa = 3.73FDD2216 pKa = 6.02GDD2218 pKa = 3.53GQADD2222 pKa = 3.82DD2223 pKa = 3.65GRR2225 pKa = 11.84YY2226 pKa = 7.14TISYY2230 pKa = 9.16AFANRR2235 pKa = 11.84RR2236 pKa = 11.84VIQPRR2241 pKa = 11.84LTVVILDD2248 pKa = 3.77YY2249 pKa = 11.59GCDD2252 pKa = 3.47LRR2254 pKa = 11.84VDD2256 pKa = 3.66QVEE2259 pKa = 4.36VTKK2262 pKa = 11.08
Molecular weight: 234.9 kDa
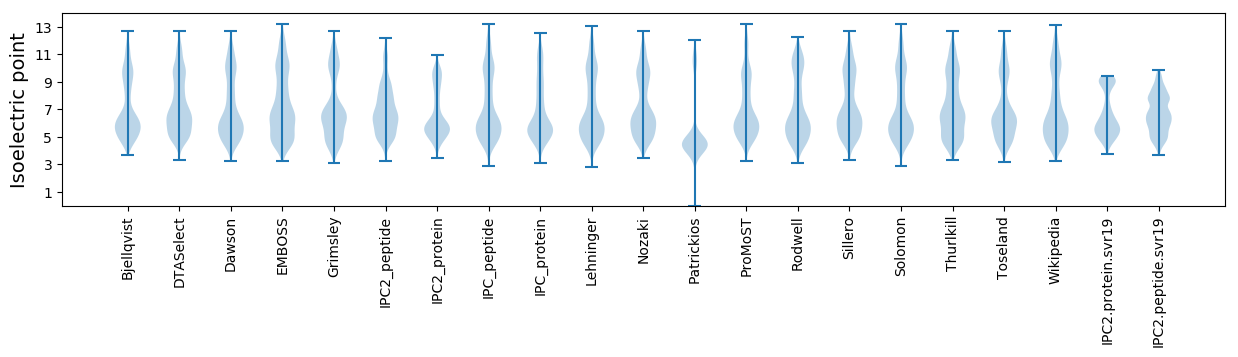
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2X3KK17|A0A2X3KK17_9BACT Probable membrane transporter protein OS=Candidatus Bipolaricaulis anaerobius OX=2026885 GN=BARAN1_0738 PE=3 SV=1
MM1 pKa = 7.4VFFLLWPIVLIPIGLALRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84FTRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 3.37
MM1 pKa = 7.4VFFLLWPIVLIPIGLALRR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84FTRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 3.37
Molecular weight: 3.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
415242 |
20 |
2262 |
314.8 |
34.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.79 ± 0.126 | 0.935 ± 0.025 |
4.875 ± 0.068 | 6.77 ± 0.071 |
3.353 ± 0.04 | 9.494 ± 0.07 |
2.023 ± 0.031 | 4.407 ± 0.043 |
2.468 ± 0.046 | 11.225 ± 0.097 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.777 ± 0.029 | 1.734 ± 0.034 |
5.953 ± 0.052 | 2.649 ± 0.035 |
8.197 ± 0.093 | 4.45 ± 0.053 |
5.141 ± 0.081 | 8.818 ± 0.06 |
1.529 ± 0.031 | 2.415 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
