
Streptomyces phage Yaboi
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Karimacvirus; Streptomyces virus Yaboi
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
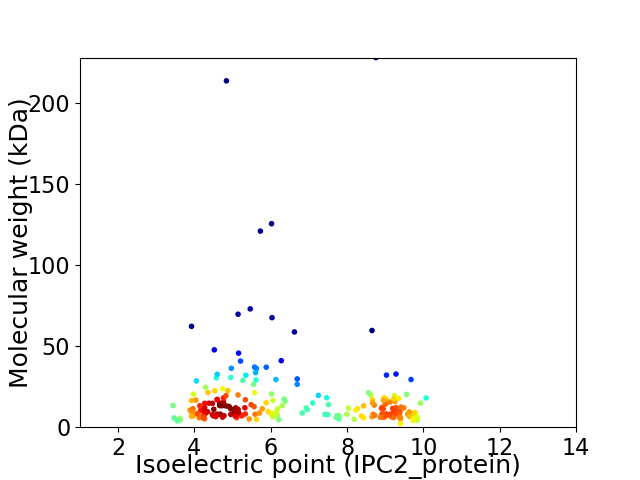
Virtual 2D-PAGE plot for 215 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A385UIB5|A0A385UIB5_9CAUD Deoxycytidylate deaminase OS=Streptomyces phage Yaboi OX=2301621 GN=233 PE=4 SV=1
MM1 pKa = 7.76SYY3 pKa = 10.54QLQVLADD10 pKa = 3.8SPFSYY15 pKa = 9.85WKK17 pKa = 10.4LDD19 pKa = 3.39GSGPVYY25 pKa = 10.65NDD27 pKa = 2.88SAGSLRR33 pKa = 11.84QADD36 pKa = 4.11LTSTAVIHH44 pKa = 6.43PALILGSGNALVLQNTNQLQMDD66 pKa = 4.02DD67 pKa = 3.63PVFNKK72 pKa = 10.33GYY74 pKa = 10.08EE75 pKa = 4.01SRR77 pKa = 11.84QFSLEE82 pKa = 3.7AWVKK86 pKa = 8.87PLSITGEE93 pKa = 4.05VSIMSHH99 pKa = 5.47NEE101 pKa = 3.62NYY103 pKa = 10.35DD104 pKa = 3.35GLTITPNRR112 pKa = 11.84IYY114 pKa = 10.76FRR116 pKa = 11.84TKK118 pKa = 10.47YY119 pKa = 8.76LTAPASEE126 pKa = 3.96ISYY129 pKa = 10.46KK130 pKa = 10.56YY131 pKa = 9.06DD132 pKa = 3.1TAKK135 pKa = 10.56AFHH138 pKa = 6.23VVGVHH143 pKa = 5.22TNGKK147 pKa = 8.05NALYY151 pKa = 10.85VDD153 pKa = 4.22GQLVAEE159 pKa = 4.57IDD161 pKa = 3.52ITDD164 pKa = 3.79EE165 pKa = 4.18QSADD169 pKa = 2.98AYY171 pKa = 11.21NFVSSDD177 pKa = 4.17LIAGQSATSSAIALDD192 pKa = 3.69APAIYY197 pKa = 9.9AAPLSAEE204 pKa = 4.46TIKK207 pKa = 10.61KK208 pKa = 9.81HH209 pKa = 5.2YY210 pKa = 10.31DD211 pKa = 3.12YY212 pKa = 11.25GIDD215 pKa = 3.73VEE217 pKa = 4.48SSEE220 pKa = 5.77AIAGYY225 pKa = 10.78NDD227 pKa = 3.2ASHH230 pKa = 6.97WNFADD235 pKa = 3.4SARR238 pKa = 11.84NIAVSKK244 pKa = 10.59LWDD247 pKa = 3.82DD248 pKa = 4.39EE249 pKa = 4.18EE250 pKa = 5.09WIDD253 pKa = 5.69GIMTDD258 pKa = 3.98VVIANDD264 pKa = 4.23IIVPSYY270 pKa = 10.63VQTEE274 pKa = 4.3TEE276 pKa = 4.07EE277 pKa = 4.48VIDD280 pKa = 3.99GLIVPVYY287 pKa = 10.06TNTSLPGVWTGSMEE301 pKa = 3.98IGEE304 pKa = 4.94SISNITDD311 pKa = 2.98AVIRR315 pKa = 11.84YY316 pKa = 9.0RR317 pKa = 11.84GEE319 pKa = 3.81GSFTIEE325 pKa = 4.16CSIDD329 pKa = 4.96NGDD332 pKa = 3.06TWQSVDD338 pKa = 3.54SAPGFSLTTEE348 pKa = 3.91DD349 pKa = 4.18SILVRR354 pKa = 11.84VTFAGGIVDD363 pKa = 3.98DD364 pKa = 4.16QSYY367 pKa = 10.9VDD369 pKa = 4.38WIQIVAYY376 pKa = 9.33SDD378 pKa = 3.43KK379 pKa = 10.75VFNGTRR385 pKa = 11.84SDD387 pKa = 3.58RR388 pKa = 11.84QATLTGTAVMARR400 pKa = 11.84EE401 pKa = 3.91FFEE404 pKa = 5.04PIEE407 pKa = 4.06YY408 pKa = 10.78ADD410 pKa = 4.32DD411 pKa = 3.85NGIKK415 pKa = 10.06ILTGDD420 pKa = 3.15ISVGVDD426 pKa = 2.87SSYY429 pKa = 11.59DD430 pKa = 3.59GEE432 pKa = 4.39EE433 pKa = 3.84EE434 pKa = 5.31AGDD437 pKa = 4.0LNINGIDD444 pKa = 3.24IWIKK448 pKa = 9.89PVAGNIITAGSASISRR464 pKa = 11.84VGNTIVFSGFSDD476 pKa = 3.79VVVNGVPVTSGTTVFTTDD494 pKa = 1.94SWYY497 pKa = 10.62HH498 pKa = 4.79IAGVFTTPGNYY509 pKa = 10.25ALTMGVANARR519 pKa = 11.84IAQASAIYY527 pKa = 10.65GDD529 pKa = 3.94IDD531 pKa = 4.94LDD533 pKa = 5.07GIQQMYY539 pKa = 9.75SAYY542 pKa = 10.55LGLPGLTVDD551 pKa = 4.56DD552 pKa = 4.2VTPIGIDD559 pKa = 3.27TANPEE564 pKa = 4.32ALLYY568 pKa = 10.09AHH570 pKa = 6.76VWSISPAGG578 pKa = 3.42
MM1 pKa = 7.76SYY3 pKa = 10.54QLQVLADD10 pKa = 3.8SPFSYY15 pKa = 9.85WKK17 pKa = 10.4LDD19 pKa = 3.39GSGPVYY25 pKa = 10.65NDD27 pKa = 2.88SAGSLRR33 pKa = 11.84QADD36 pKa = 4.11LTSTAVIHH44 pKa = 6.43PALILGSGNALVLQNTNQLQMDD66 pKa = 4.02DD67 pKa = 3.63PVFNKK72 pKa = 10.33GYY74 pKa = 10.08EE75 pKa = 4.01SRR77 pKa = 11.84QFSLEE82 pKa = 3.7AWVKK86 pKa = 8.87PLSITGEE93 pKa = 4.05VSIMSHH99 pKa = 5.47NEE101 pKa = 3.62NYY103 pKa = 10.35DD104 pKa = 3.35GLTITPNRR112 pKa = 11.84IYY114 pKa = 10.76FRR116 pKa = 11.84TKK118 pKa = 10.47YY119 pKa = 8.76LTAPASEE126 pKa = 3.96ISYY129 pKa = 10.46KK130 pKa = 10.56YY131 pKa = 9.06DD132 pKa = 3.1TAKK135 pKa = 10.56AFHH138 pKa = 6.23VVGVHH143 pKa = 5.22TNGKK147 pKa = 8.05NALYY151 pKa = 10.85VDD153 pKa = 4.22GQLVAEE159 pKa = 4.57IDD161 pKa = 3.52ITDD164 pKa = 3.79EE165 pKa = 4.18QSADD169 pKa = 2.98AYY171 pKa = 11.21NFVSSDD177 pKa = 4.17LIAGQSATSSAIALDD192 pKa = 3.69APAIYY197 pKa = 9.9AAPLSAEE204 pKa = 4.46TIKK207 pKa = 10.61KK208 pKa = 9.81HH209 pKa = 5.2YY210 pKa = 10.31DD211 pKa = 3.12YY212 pKa = 11.25GIDD215 pKa = 3.73VEE217 pKa = 4.48SSEE220 pKa = 5.77AIAGYY225 pKa = 10.78NDD227 pKa = 3.2ASHH230 pKa = 6.97WNFADD235 pKa = 3.4SARR238 pKa = 11.84NIAVSKK244 pKa = 10.59LWDD247 pKa = 3.82DD248 pKa = 4.39EE249 pKa = 4.18EE250 pKa = 5.09WIDD253 pKa = 5.69GIMTDD258 pKa = 3.98VVIANDD264 pKa = 4.23IIVPSYY270 pKa = 10.63VQTEE274 pKa = 4.3TEE276 pKa = 4.07EE277 pKa = 4.48VIDD280 pKa = 3.99GLIVPVYY287 pKa = 10.06TNTSLPGVWTGSMEE301 pKa = 3.98IGEE304 pKa = 4.94SISNITDD311 pKa = 2.98AVIRR315 pKa = 11.84YY316 pKa = 9.0RR317 pKa = 11.84GEE319 pKa = 3.81GSFTIEE325 pKa = 4.16CSIDD329 pKa = 4.96NGDD332 pKa = 3.06TWQSVDD338 pKa = 3.54SAPGFSLTTEE348 pKa = 3.91DD349 pKa = 4.18SILVRR354 pKa = 11.84VTFAGGIVDD363 pKa = 3.98DD364 pKa = 4.16QSYY367 pKa = 10.9VDD369 pKa = 4.38WIQIVAYY376 pKa = 9.33SDD378 pKa = 3.43KK379 pKa = 10.75VFNGTRR385 pKa = 11.84SDD387 pKa = 3.58RR388 pKa = 11.84QATLTGTAVMARR400 pKa = 11.84EE401 pKa = 3.91FFEE404 pKa = 5.04PIEE407 pKa = 4.06YY408 pKa = 10.78ADD410 pKa = 4.32DD411 pKa = 3.85NGIKK415 pKa = 10.06ILTGDD420 pKa = 3.15ISVGVDD426 pKa = 2.87SSYY429 pKa = 11.59DD430 pKa = 3.59GEE432 pKa = 4.39EE433 pKa = 3.84EE434 pKa = 5.31AGDD437 pKa = 4.0LNINGIDD444 pKa = 3.24IWIKK448 pKa = 9.89PVAGNIITAGSASISRR464 pKa = 11.84VGNTIVFSGFSDD476 pKa = 3.79VVVNGVPVTSGTTVFTTDD494 pKa = 1.94SWYY497 pKa = 10.62HH498 pKa = 4.79IAGVFTTPGNYY509 pKa = 10.25ALTMGVANARR519 pKa = 11.84IAQASAIYY527 pKa = 10.65GDD529 pKa = 3.94IDD531 pKa = 4.94LDD533 pKa = 5.07GIQQMYY539 pKa = 9.75SAYY542 pKa = 10.55LGLPGLTVDD551 pKa = 4.56DD552 pKa = 4.2VTPIGIDD559 pKa = 3.27TANPEE564 pKa = 4.32ALLYY568 pKa = 10.09AHH570 pKa = 6.76VWSISPAGG578 pKa = 3.42
Molecular weight: 62.14 kDa
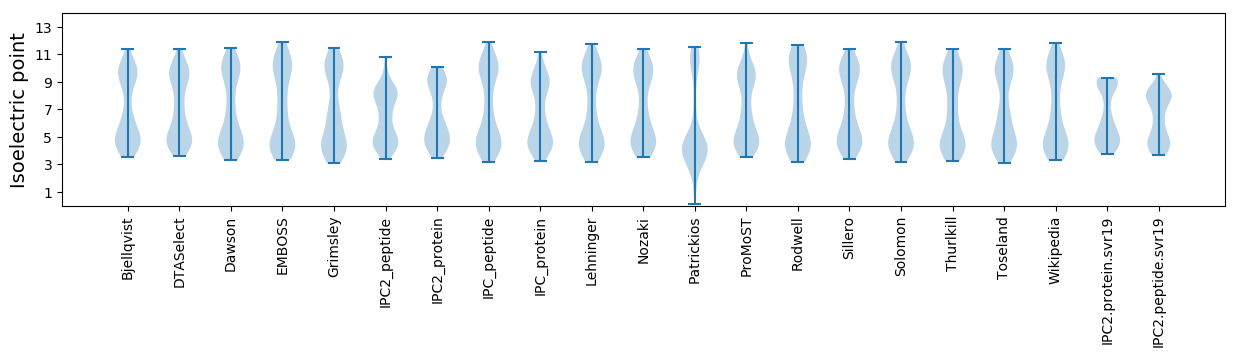
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A385UH49|A0A385UH49_9CAUD Uncharacterized protein OS=Streptomyces phage Yaboi OX=2301621 GN=135 PE=4 SV=1
MM1 pKa = 7.34TVTLLSRR8 pKa = 11.84RR9 pKa = 11.84LSRR12 pKa = 11.84CHH14 pKa = 5.7TLRR17 pKa = 11.84NVTVSLCHH25 pKa = 6.06ICHH28 pKa = 6.79FLICHH33 pKa = 6.37APGGGRR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84GSAFVVLYY49 pKa = 9.78ILRR52 pKa = 11.84RR53 pKa = 11.84SKK55 pKa = 11.09GRR57 pKa = 11.84GDD59 pKa = 3.2LTEE62 pKa = 3.64QEE64 pKa = 4.52RR65 pKa = 11.84PVKK68 pKa = 10.57VNYY71 pKa = 8.23TGKK74 pKa = 10.33RR75 pKa = 11.84KK76 pKa = 9.71EE77 pKa = 4.04KK78 pKa = 10.28EE79 pKa = 3.73
MM1 pKa = 7.34TVTLLSRR8 pKa = 11.84RR9 pKa = 11.84LSRR12 pKa = 11.84CHH14 pKa = 5.7TLRR17 pKa = 11.84NVTVSLCHH25 pKa = 6.06ICHH28 pKa = 6.79FLICHH33 pKa = 6.37APGGGRR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84GSAFVVLYY49 pKa = 9.78ILRR52 pKa = 11.84RR53 pKa = 11.84SKK55 pKa = 11.09GRR57 pKa = 11.84GDD59 pKa = 3.2LTEE62 pKa = 3.64QEE64 pKa = 4.52RR65 pKa = 11.84PVKK68 pKa = 10.57VNYY71 pKa = 8.23TGKK74 pKa = 10.33RR75 pKa = 11.84KK76 pKa = 9.71EE77 pKa = 4.04KK78 pKa = 10.28EE79 pKa = 3.73
Molecular weight: 9.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
33893 |
20 |
2093 |
157.6 |
17.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.258 ± 0.366 | 1.083 ± 0.134 |
6.379 ± 0.201 | 7.173 ± 0.364 |
3.809 ± 0.123 | 7.373 ± 0.175 |
1.729 ± 0.121 | 5.497 ± 0.149 |
6.497 ± 0.342 | 7.305 ± 0.211 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.915 ± 0.137 | 4.682 ± 0.158 |
3.65 ± 0.166 | 3.204 ± 0.187 |
5.73 ± 0.196 | 5.957 ± 0.294 |
6.01 ± 0.394 | 7.099 ± 0.228 |
1.947 ± 0.095 | 3.703 ± 0.167 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
