
Streptococcus satellite phage Javan593
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.83
Get precalculated fractions of proteins
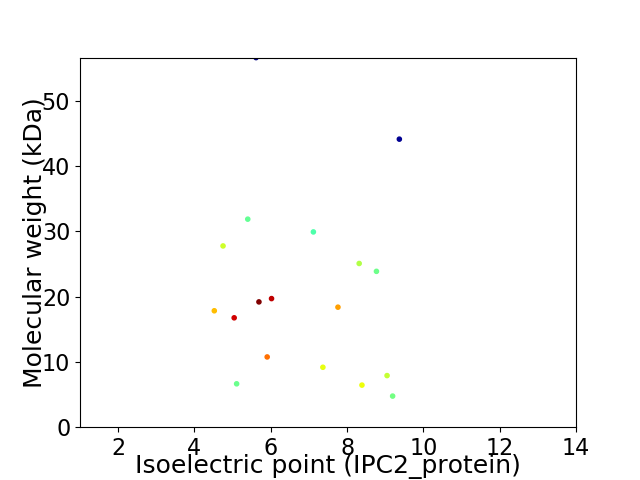
Virtual 2D-PAGE plot for 18 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
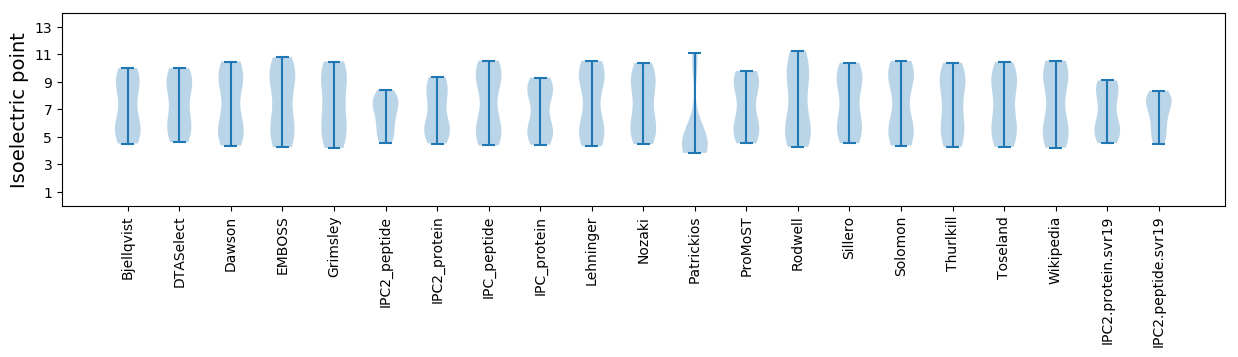
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D5ZTQ6|A0A4D5ZTQ6_9VIRU Uncharacterized protein OS=Streptococcus satellite phage Javan593 OX=2558763 GN=JavanS593_0016 PE=4 SV=1
MM1 pKa = 7.56TEE3 pKa = 3.71QEE5 pKa = 4.16YY6 pKa = 9.56FAQAEE11 pKa = 4.43KK12 pKa = 10.67EE13 pKa = 4.12LEE15 pKa = 3.97EE16 pKa = 5.29LNQQRR21 pKa = 11.84ADD23 pKa = 3.91FMAMDD28 pKa = 4.34FEE30 pKa = 4.67EE31 pKa = 5.98LNTADD36 pKa = 4.21YY37 pKa = 11.27INFLEE42 pKa = 4.21IGNRR46 pKa = 11.84IVAEE50 pKa = 4.1DD51 pKa = 3.43VTLNVYY57 pKa = 9.26EE58 pKa = 4.74LYY60 pKa = 10.55KK61 pKa = 11.05HH62 pKa = 6.84PDD64 pKa = 2.79TRR66 pKa = 11.84AKK68 pKa = 10.46FFATIAKK75 pKa = 9.47IAYY78 pKa = 8.24HH79 pKa = 5.81VNNMFQTADD88 pKa = 3.41RR89 pKa = 11.84MEE91 pKa = 4.33AMIDD95 pKa = 3.46SLEE98 pKa = 3.8QHH100 pKa = 5.84YY101 pKa = 11.25QNTIKK106 pKa = 10.85KK107 pKa = 9.71LVYY110 pKa = 9.26QTDD113 pKa = 3.56SDD115 pKa = 4.12KK116 pKa = 11.29LAEE119 pKa = 4.57LLLEE123 pKa = 4.38VKK125 pKa = 10.18KK126 pKa = 10.9DD127 pKa = 3.87NPNMTAEE134 pKa = 4.19QEE136 pKa = 4.42SQFIRR141 pKa = 11.84DD142 pKa = 3.67VAVSGLLAKK151 pKa = 10.56EE152 pKa = 4.02NN153 pKa = 3.7
MM1 pKa = 7.56TEE3 pKa = 3.71QEE5 pKa = 4.16YY6 pKa = 9.56FAQAEE11 pKa = 4.43KK12 pKa = 10.67EE13 pKa = 4.12LEE15 pKa = 3.97EE16 pKa = 5.29LNQQRR21 pKa = 11.84ADD23 pKa = 3.91FMAMDD28 pKa = 4.34FEE30 pKa = 4.67EE31 pKa = 5.98LNTADD36 pKa = 4.21YY37 pKa = 11.27INFLEE42 pKa = 4.21IGNRR46 pKa = 11.84IVAEE50 pKa = 4.1DD51 pKa = 3.43VTLNVYY57 pKa = 9.26EE58 pKa = 4.74LYY60 pKa = 10.55KK61 pKa = 11.05HH62 pKa = 6.84PDD64 pKa = 2.79TRR66 pKa = 11.84AKK68 pKa = 10.46FFATIAKK75 pKa = 9.47IAYY78 pKa = 8.24HH79 pKa = 5.81VNNMFQTADD88 pKa = 3.41RR89 pKa = 11.84MEE91 pKa = 4.33AMIDD95 pKa = 3.46SLEE98 pKa = 3.8QHH100 pKa = 5.84YY101 pKa = 11.25QNTIKK106 pKa = 10.85KK107 pKa = 9.71LVYY110 pKa = 9.26QTDD113 pKa = 3.56SDD115 pKa = 4.12KK116 pKa = 11.29LAEE119 pKa = 4.57LLLEE123 pKa = 4.38VKK125 pKa = 10.18KK126 pKa = 10.9DD127 pKa = 3.87NPNMTAEE134 pKa = 4.19QEE136 pKa = 4.42SQFIRR141 pKa = 11.84DD142 pKa = 3.67VAVSGLLAKK151 pKa = 10.56EE152 pKa = 4.02NN153 pKa = 3.7
Molecular weight: 17.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D5ZWD1|A0A4D5ZWD1_9VIRU Integrase OS=Streptococcus satellite phage Javan593 OX=2558763 GN=JavanS593_0017 PE=3 SV=1
MM1 pKa = 7.65KK2 pKa = 10.14IKK4 pKa = 10.43EE5 pKa = 4.14VTKK8 pKa = 10.29KK9 pKa = 10.58DD10 pKa = 3.16GSIVYY15 pKa = 10.15RR16 pKa = 11.84SNIYY20 pKa = 10.61LGVDD24 pKa = 3.26QITGKK29 pKa = 10.2KK30 pKa = 9.93VKK32 pKa = 9.98TNVTGRR38 pKa = 11.84TQKK41 pKa = 10.12EE42 pKa = 3.98VRR44 pKa = 11.84QKK46 pKa = 11.28AKK48 pKa = 9.23QAEE51 pKa = 4.54YY52 pKa = 10.95NFKK55 pKa = 10.73QSGSTRR61 pKa = 11.84FCAQTDD67 pKa = 2.98IKK69 pKa = 10.02TIEE72 pKa = 4.23EE73 pKa = 4.36LSEE76 pKa = 3.9SWLANYY82 pKa = 9.94QNTVKK87 pKa = 10.37PQTLRR92 pKa = 11.84NTKK95 pKa = 10.24SYY97 pKa = 10.52LKK99 pKa = 10.53NHH101 pKa = 6.59ILPRR105 pKa = 11.84LGDD108 pKa = 3.47MRR110 pKa = 11.84LEE112 pKa = 4.2KK113 pKa = 10.58LSPPVAQAFINDD125 pKa = 3.68LAKK128 pKa = 9.9RR129 pKa = 11.84TNQFDD134 pKa = 3.59KK135 pKa = 11.02ARR137 pKa = 11.84SVLSRR142 pKa = 11.84MFRR145 pKa = 11.84YY146 pKa = 10.18AIVLQLVQYY155 pKa = 10.86NPVRR159 pKa = 11.84DD160 pKa = 3.64TLVPRR165 pKa = 11.84KK166 pKa = 9.55KK167 pKa = 10.18QSAKK171 pKa = 10.62KK172 pKa = 8.61KK173 pKa = 9.34VKK175 pKa = 10.38YY176 pKa = 9.74IQSDD180 pKa = 3.63HH181 pKa = 6.4LKK183 pKa = 10.68KK184 pKa = 10.75FLDD187 pKa = 5.06HH188 pKa = 7.05IDD190 pKa = 3.63KK191 pKa = 10.65LAQKK195 pKa = 10.73DD196 pKa = 3.84FTRR199 pKa = 11.84YY200 pKa = 9.52GYY202 pKa = 9.19MVAFRR207 pKa = 11.84LLLATGMRR215 pKa = 11.84IGEE218 pKa = 4.32LSALEE223 pKa = 3.83WSDD226 pKa = 3.25INLEE230 pKa = 4.02EE231 pKa = 4.42HH232 pKa = 7.15SIRR235 pKa = 11.84INKK238 pKa = 8.86TYY240 pKa = 10.28LQEE243 pKa = 4.02IKK245 pKa = 10.5QVGEE249 pKa = 4.25TKK251 pKa = 9.8TKK253 pKa = 9.49AGEE256 pKa = 4.08RR257 pKa = 11.84VVSIDD262 pKa = 3.05KK263 pKa = 9.71ATVLMLKK270 pKa = 9.98QYY272 pKa = 8.73QNRR275 pKa = 11.84QRR277 pKa = 11.84VVFLEE282 pKa = 4.08VGGAPTRR289 pKa = 11.84VFEE292 pKa = 4.45TPTRR296 pKa = 11.84LYY298 pKa = 9.59LTRR301 pKa = 11.84NNFQRR306 pKa = 11.84VLDD309 pKa = 3.66HH310 pKa = 6.85HH311 pKa = 6.68CEE313 pKa = 3.79KK314 pKa = 11.27LEE316 pKa = 3.84IPRR319 pKa = 11.84VTLHH323 pKa = 7.05AFRR326 pKa = 11.84HH327 pKa = 4.68THH329 pKa = 7.12ASLLLNAGISYY340 pKa = 10.65KK341 pKa = 10.3EE342 pKa = 3.81LQHH345 pKa = 7.18RR346 pKa = 11.84LGHH349 pKa = 5.94SNITLTLDD357 pKa = 3.69TYY359 pKa = 11.66SHH361 pKa = 7.12LSKK364 pKa = 10.9DD365 pKa = 3.57KK366 pKa = 10.41EE367 pKa = 4.25KK368 pKa = 10.93EE369 pKa = 3.85AVSYY373 pKa = 9.71YY374 pKa = 10.5EE375 pKa = 4.33KK376 pKa = 11.08ALASLL381 pKa = 4.18
MM1 pKa = 7.65KK2 pKa = 10.14IKK4 pKa = 10.43EE5 pKa = 4.14VTKK8 pKa = 10.29KK9 pKa = 10.58DD10 pKa = 3.16GSIVYY15 pKa = 10.15RR16 pKa = 11.84SNIYY20 pKa = 10.61LGVDD24 pKa = 3.26QITGKK29 pKa = 10.2KK30 pKa = 9.93VKK32 pKa = 9.98TNVTGRR38 pKa = 11.84TQKK41 pKa = 10.12EE42 pKa = 3.98VRR44 pKa = 11.84QKK46 pKa = 11.28AKK48 pKa = 9.23QAEE51 pKa = 4.54YY52 pKa = 10.95NFKK55 pKa = 10.73QSGSTRR61 pKa = 11.84FCAQTDD67 pKa = 2.98IKK69 pKa = 10.02TIEE72 pKa = 4.23EE73 pKa = 4.36LSEE76 pKa = 3.9SWLANYY82 pKa = 9.94QNTVKK87 pKa = 10.37PQTLRR92 pKa = 11.84NTKK95 pKa = 10.24SYY97 pKa = 10.52LKK99 pKa = 10.53NHH101 pKa = 6.59ILPRR105 pKa = 11.84LGDD108 pKa = 3.47MRR110 pKa = 11.84LEE112 pKa = 4.2KK113 pKa = 10.58LSPPVAQAFINDD125 pKa = 3.68LAKK128 pKa = 9.9RR129 pKa = 11.84TNQFDD134 pKa = 3.59KK135 pKa = 11.02ARR137 pKa = 11.84SVLSRR142 pKa = 11.84MFRR145 pKa = 11.84YY146 pKa = 10.18AIVLQLVQYY155 pKa = 10.86NPVRR159 pKa = 11.84DD160 pKa = 3.64TLVPRR165 pKa = 11.84KK166 pKa = 9.55KK167 pKa = 10.18QSAKK171 pKa = 10.62KK172 pKa = 8.61KK173 pKa = 9.34VKK175 pKa = 10.38YY176 pKa = 9.74IQSDD180 pKa = 3.63HH181 pKa = 6.4LKK183 pKa = 10.68KK184 pKa = 10.75FLDD187 pKa = 5.06HH188 pKa = 7.05IDD190 pKa = 3.63KK191 pKa = 10.65LAQKK195 pKa = 10.73DD196 pKa = 3.84FTRR199 pKa = 11.84YY200 pKa = 9.52GYY202 pKa = 9.19MVAFRR207 pKa = 11.84LLLATGMRR215 pKa = 11.84IGEE218 pKa = 4.32LSALEE223 pKa = 3.83WSDD226 pKa = 3.25INLEE230 pKa = 4.02EE231 pKa = 4.42HH232 pKa = 7.15SIRR235 pKa = 11.84INKK238 pKa = 8.86TYY240 pKa = 10.28LQEE243 pKa = 4.02IKK245 pKa = 10.5QVGEE249 pKa = 4.25TKK251 pKa = 9.8TKK253 pKa = 9.49AGEE256 pKa = 4.08RR257 pKa = 11.84VVSIDD262 pKa = 3.05KK263 pKa = 9.71ATVLMLKK270 pKa = 9.98QYY272 pKa = 8.73QNRR275 pKa = 11.84QRR277 pKa = 11.84VVFLEE282 pKa = 4.08VGGAPTRR289 pKa = 11.84VFEE292 pKa = 4.45TPTRR296 pKa = 11.84LYY298 pKa = 9.59LTRR301 pKa = 11.84NNFQRR306 pKa = 11.84VLDD309 pKa = 3.66HH310 pKa = 6.85HH311 pKa = 6.68CEE313 pKa = 3.79KK314 pKa = 11.27LEE316 pKa = 3.84IPRR319 pKa = 11.84VTLHH323 pKa = 7.05AFRR326 pKa = 11.84HH327 pKa = 4.68THH329 pKa = 7.12ASLLLNAGISYY340 pKa = 10.65KK341 pKa = 10.3EE342 pKa = 3.81LQHH345 pKa = 7.18RR346 pKa = 11.84LGHH349 pKa = 5.94SNITLTLDD357 pKa = 3.69TYY359 pKa = 11.66SHH361 pKa = 7.12LSKK364 pKa = 10.9DD365 pKa = 3.57KK366 pKa = 10.41EE367 pKa = 4.25KK368 pKa = 10.93EE369 pKa = 3.85AVSYY373 pKa = 9.71YY374 pKa = 10.5EE375 pKa = 4.33KK376 pKa = 11.08ALASLL381 pKa = 4.18
Molecular weight: 44.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3242 |
41 |
496 |
180.1 |
20.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.151 ± 0.554 | 0.401 ± 0.144 |
5.552 ± 0.725 | 8.822 ± 0.788 |
4.257 ± 0.293 | 4.349 ± 0.34 |
2.005 ± 0.244 | 7.465 ± 0.502 |
9.469 ± 0.42 | 10.672 ± 0.443 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.283 ± 0.296 | 6.231 ± 0.498 |
2.992 ± 0.453 | 4.627 ± 0.658 |
4.935 ± 0.422 | 5.891 ± 0.396 |
5.46 ± 0.492 | 4.226 ± 0.432 |
0.802 ± 0.144 | 4.411 ± 0.321 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
