
Terriglobus albidus
Taxonomy: cellular organisms; Bacteria; Acidobacteria; Acidobacteriia; Acidobacteriales; Acidobacteriaceae; Terriglobus
Average proteome isoelectric point is 6.77
Get precalculated fractions of proteins
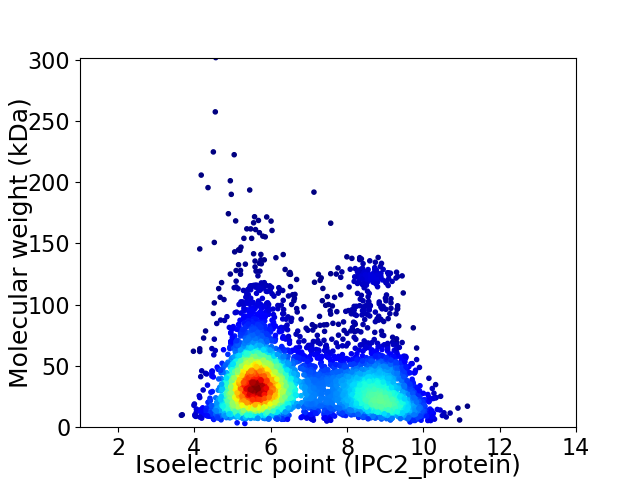
Virtual 2D-PAGE plot for 4970 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5B9EGS6|A0A5B9EGS6_9BACT Glycerophosphodiester phosphodiesterase OS=Terriglobus albidus OX=1592106 GN=FTW19_21595 PE=4 SV=1
MM1 pKa = 8.02RR2 pKa = 11.84FPFVPVLLLLASSSILPAQTNLSFEE27 pKa = 4.62LSRR30 pKa = 11.84DD31 pKa = 3.1TYY33 pKa = 10.07MAFGVEE39 pKa = 4.98AMAQGDD45 pKa = 4.07FNGDD49 pKa = 3.51GKK51 pKa = 10.23PDD53 pKa = 3.47MVFGGGSSFTVVTLRR68 pKa = 11.84LGNGDD73 pKa = 3.66GTFQPPITVGQADD86 pKa = 3.62SSEE89 pKa = 4.54VEE91 pKa = 4.87DD92 pKa = 5.04IAAADD97 pKa = 3.99LNQDD101 pKa = 3.1GKK103 pKa = 11.47LDD105 pKa = 4.19VIVLCIGGTFDD116 pKa = 4.19VFLGNGDD123 pKa = 4.08GSFQPAIAVATAGSPRR139 pKa = 11.84AMAVGDD145 pKa = 3.74FNGDD149 pKa = 3.35QRR151 pKa = 11.84PDD153 pKa = 3.32LAIGDD158 pKa = 4.18SQGSVEE164 pKa = 4.82IFNNTDD170 pKa = 2.46GKK172 pKa = 10.72NFVLSNTVPIDD183 pKa = 3.36PTRR186 pKa = 11.84DD187 pKa = 3.16ILNVKK192 pKa = 10.37AGDD195 pKa = 3.79FDD197 pKa = 6.21DD198 pKa = 6.49DD199 pKa = 4.23GVTNLAVLTSDD210 pKa = 4.12SAYY213 pKa = 10.86ALWNDD218 pKa = 3.41GAGNFRR224 pKa = 11.84KK225 pKa = 9.82SALSAYY231 pKa = 7.36TNPAGLNVGDD241 pKa = 4.69LNQDD245 pKa = 2.83GMTDD249 pKa = 2.98ILVSYY254 pKa = 9.63DD255 pKa = 3.41CHH257 pKa = 7.09PNPQYY262 pKa = 10.17PGPKK266 pKa = 10.06GPISTCEE273 pKa = 3.98GVDD276 pKa = 3.26VFYY279 pKa = 10.88GQGQQKK285 pKa = 7.79TFEE288 pKa = 4.12RR289 pKa = 11.84HH290 pKa = 5.83IITADD295 pKa = 3.19TVGAARR301 pKa = 11.84YY302 pKa = 8.47LWAADD307 pKa = 3.61VNGDD311 pKa = 4.11GIGDD315 pKa = 4.64LVTGTSDD322 pKa = 3.64QNGSQTGLFIWLGHH336 pKa = 6.82PDD338 pKa = 3.38GSFDD342 pKa = 3.39QAASRR347 pKa = 11.84FIATSSGTGQLVVADD362 pKa = 4.81FNRR365 pKa = 11.84DD366 pKa = 3.03GMIDD370 pKa = 4.09FAQALPGDD378 pKa = 4.43AQTQIYY384 pKa = 10.49LNATNRR390 pKa = 11.84APCATSQINPTVTVCQPVDD409 pKa = 3.6NTYY412 pKa = 11.08LPAPAIRR419 pKa = 11.84VTANAYY425 pKa = 7.85DD426 pKa = 4.14TNQVTAMQLYY436 pKa = 9.42VNGDD440 pKa = 3.66LVYY443 pKa = 10.53SRR445 pKa = 11.84PVSGFNRR452 pKa = 11.84TFQLGEE458 pKa = 3.84GSFNLVTKK466 pKa = 10.17AWDD469 pKa = 3.57YY470 pKa = 11.69AGLNFRR476 pKa = 11.84SVRR479 pKa = 11.84HH480 pKa = 3.9VTLYY484 pKa = 10.8NGTPGSVCAAALGTAEE500 pKa = 3.95ICLPSGATSTSPVHH514 pKa = 6.31IVGNGYY520 pKa = 7.88TSYY523 pKa = 11.35VPTAAQLYY531 pKa = 9.43IDD533 pKa = 4.54GNLVINNTSCNDD545 pKa = 3.28YY546 pKa = 10.22GACPGGSSLVDD557 pKa = 3.53TYY559 pKa = 11.49QGLSSGSHH567 pKa = 6.28DD568 pKa = 4.94LVFKK572 pKa = 10.94LWDD575 pKa = 3.74ANGNIYY581 pKa = 10.7SADD584 pKa = 3.25KK585 pKa = 9.88TVSVEE590 pKa = 3.82
MM1 pKa = 8.02RR2 pKa = 11.84FPFVPVLLLLASSSILPAQTNLSFEE27 pKa = 4.62LSRR30 pKa = 11.84DD31 pKa = 3.1TYY33 pKa = 10.07MAFGVEE39 pKa = 4.98AMAQGDD45 pKa = 4.07FNGDD49 pKa = 3.51GKK51 pKa = 10.23PDD53 pKa = 3.47MVFGGGSSFTVVTLRR68 pKa = 11.84LGNGDD73 pKa = 3.66GTFQPPITVGQADD86 pKa = 3.62SSEE89 pKa = 4.54VEE91 pKa = 4.87DD92 pKa = 5.04IAAADD97 pKa = 3.99LNQDD101 pKa = 3.1GKK103 pKa = 11.47LDD105 pKa = 4.19VIVLCIGGTFDD116 pKa = 4.19VFLGNGDD123 pKa = 4.08GSFQPAIAVATAGSPRR139 pKa = 11.84AMAVGDD145 pKa = 3.74FNGDD149 pKa = 3.35QRR151 pKa = 11.84PDD153 pKa = 3.32LAIGDD158 pKa = 4.18SQGSVEE164 pKa = 4.82IFNNTDD170 pKa = 2.46GKK172 pKa = 10.72NFVLSNTVPIDD183 pKa = 3.36PTRR186 pKa = 11.84DD187 pKa = 3.16ILNVKK192 pKa = 10.37AGDD195 pKa = 3.79FDD197 pKa = 6.21DD198 pKa = 6.49DD199 pKa = 4.23GVTNLAVLTSDD210 pKa = 4.12SAYY213 pKa = 10.86ALWNDD218 pKa = 3.41GAGNFRR224 pKa = 11.84KK225 pKa = 9.82SALSAYY231 pKa = 7.36TNPAGLNVGDD241 pKa = 4.69LNQDD245 pKa = 2.83GMTDD249 pKa = 2.98ILVSYY254 pKa = 9.63DD255 pKa = 3.41CHH257 pKa = 7.09PNPQYY262 pKa = 10.17PGPKK266 pKa = 10.06GPISTCEE273 pKa = 3.98GVDD276 pKa = 3.26VFYY279 pKa = 10.88GQGQQKK285 pKa = 7.79TFEE288 pKa = 4.12RR289 pKa = 11.84HH290 pKa = 5.83IITADD295 pKa = 3.19TVGAARR301 pKa = 11.84YY302 pKa = 8.47LWAADD307 pKa = 3.61VNGDD311 pKa = 4.11GIGDD315 pKa = 4.64LVTGTSDD322 pKa = 3.64QNGSQTGLFIWLGHH336 pKa = 6.82PDD338 pKa = 3.38GSFDD342 pKa = 3.39QAASRR347 pKa = 11.84FIATSSGTGQLVVADD362 pKa = 4.81FNRR365 pKa = 11.84DD366 pKa = 3.03GMIDD370 pKa = 4.09FAQALPGDD378 pKa = 4.43AQTQIYY384 pKa = 10.49LNATNRR390 pKa = 11.84APCATSQINPTVTVCQPVDD409 pKa = 3.6NTYY412 pKa = 11.08LPAPAIRR419 pKa = 11.84VTANAYY425 pKa = 7.85DD426 pKa = 4.14TNQVTAMQLYY436 pKa = 9.42VNGDD440 pKa = 3.66LVYY443 pKa = 10.53SRR445 pKa = 11.84PVSGFNRR452 pKa = 11.84TFQLGEE458 pKa = 3.84GSFNLVTKK466 pKa = 10.17AWDD469 pKa = 3.57YY470 pKa = 11.69AGLNFRR476 pKa = 11.84SVRR479 pKa = 11.84HH480 pKa = 3.9VTLYY484 pKa = 10.8NGTPGSVCAAALGTAEE500 pKa = 3.95ICLPSGATSTSPVHH514 pKa = 6.31IVGNGYY520 pKa = 7.88TSYY523 pKa = 11.35VPTAAQLYY531 pKa = 9.43IDD533 pKa = 4.54GNLVINNTSCNDD545 pKa = 3.28YY546 pKa = 10.22GACPGGSSLVDD557 pKa = 3.53TYY559 pKa = 11.49QGLSSGSHH567 pKa = 6.28DD568 pKa = 4.94LVFKK572 pKa = 10.94LWDD575 pKa = 3.74ANGNIYY581 pKa = 10.7SADD584 pKa = 3.25KK585 pKa = 9.88TVSVEE590 pKa = 3.82
Molecular weight: 61.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5B9E5V1|A0A5B9E5V1_9BACT Ribonuclease J OS=Terriglobus albidus OX=1592106 GN=rnj PE=3 SV=1
MM1 pKa = 8.0PKK3 pKa = 9.02RR4 pKa = 11.84TFQPNRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84SKK15 pKa = 10.13VHH17 pKa = 5.86GFLSRR22 pKa = 11.84MASKK26 pKa = 10.6AGRR29 pKa = 11.84AVLSRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.19GRR40 pKa = 11.84HH41 pKa = 5.72KK42 pKa = 10.29IAVSAGFRR50 pKa = 11.84DD51 pKa = 3.66
MM1 pKa = 8.0PKK3 pKa = 9.02RR4 pKa = 11.84TFQPNRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84SKK15 pKa = 10.13VHH17 pKa = 5.86GFLSRR22 pKa = 11.84MASKK26 pKa = 10.6AGRR29 pKa = 11.84AVLSRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.19GRR40 pKa = 11.84HH41 pKa = 5.72KK42 pKa = 10.29IAVSAGFRR50 pKa = 11.84DD51 pKa = 3.66
Molecular weight: 5.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1854419 |
26 |
3063 |
373.1 |
40.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.626 ± 0.038 | 0.838 ± 0.01 |
4.963 ± 0.03 | 5.351 ± 0.044 |
3.916 ± 0.02 | 8.015 ± 0.037 |
2.249 ± 0.017 | 5.046 ± 0.024 |
3.758 ± 0.031 | 9.915 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.271 ± 0.016 | 3.511 ± 0.039 |
5.248 ± 0.022 | 3.986 ± 0.024 |
6.157 ± 0.039 | 6.446 ± 0.041 |
6.243 ± 0.049 | 7.204 ± 0.029 |
1.417 ± 0.015 | 2.841 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
