
Calla lily chlorotic spot virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Tospoviridae; Orthotospovirus; Calla lily chlorotic spot orthotospovirus
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
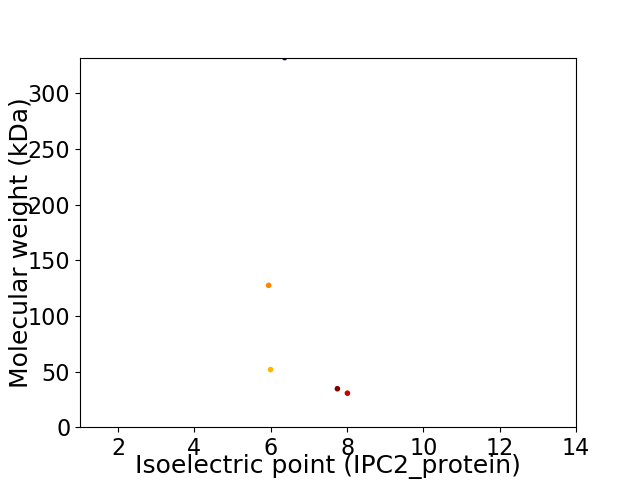
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
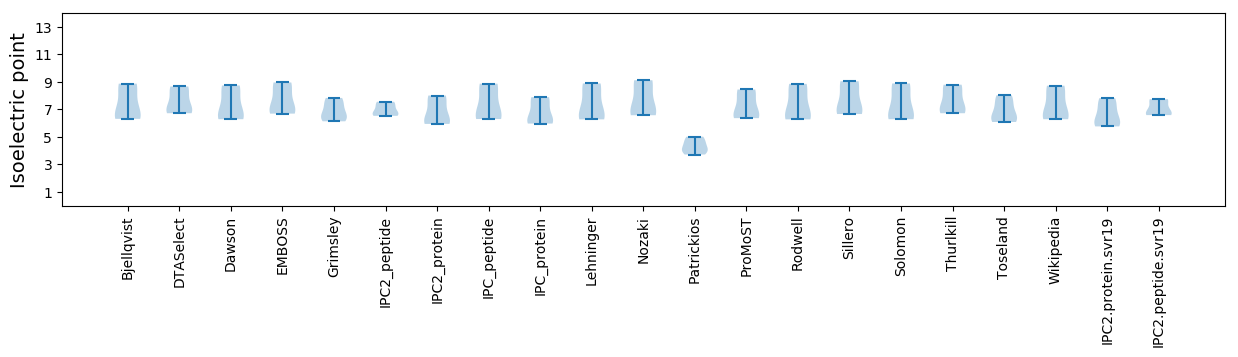
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H4WN56|A0A2H4WN56_9VIRU Nucleoprotein OS=Calla lily chlorotic spot virus OX=309542 PE=3 SV=1
MM1 pKa = 7.43KK2 pKa = 10.11KK3 pKa = 10.44YY4 pKa = 10.55YY5 pKa = 10.58LPLYY9 pKa = 9.91CLSLLLLFFVSEE21 pKa = 4.29VFLLSKK27 pKa = 9.69TEE29 pKa = 3.94HH30 pKa = 6.97DD31 pKa = 4.1SEE33 pKa = 4.03ITRR36 pKa = 11.84IRR38 pKa = 11.84DD39 pKa = 3.51RR40 pKa = 11.84YY41 pKa = 9.72PVDD44 pKa = 4.83DD45 pKa = 5.4PDD47 pKa = 5.87DD48 pKa = 3.92LVEE51 pKa = 4.94NYY53 pKa = 10.76GEE55 pKa = 4.46YY56 pKa = 9.98IPHH59 pKa = 6.53RR60 pKa = 11.84TPEE63 pKa = 3.96ADD65 pKa = 3.43KK66 pKa = 8.95PTKK69 pKa = 10.02VLSRR73 pKa = 11.84ILRR76 pKa = 11.84DD77 pKa = 3.46SSSTEE82 pKa = 3.6APRR85 pKa = 11.84LDD87 pKa = 4.97SFDD90 pKa = 3.49CMKK93 pKa = 10.53FEE95 pKa = 5.84KK96 pKa = 9.09KK97 pKa = 9.42QCMIKK102 pKa = 10.61GLSDD106 pKa = 3.94FNAHH110 pKa = 5.15YY111 pKa = 10.71QIDD114 pKa = 3.95NGNEE118 pKa = 3.73IVSCISDD125 pKa = 3.47SPNIFEE131 pKa = 3.85ICQYY135 pKa = 10.55EE136 pKa = 4.44KK137 pKa = 10.93EE138 pKa = 4.22FKK140 pKa = 9.05KK141 pKa = 9.42TKK143 pKa = 9.73FSKK146 pKa = 10.89LPVLPVLKK154 pKa = 10.71LEE156 pKa = 4.26NKK158 pKa = 9.53RR159 pKa = 11.84VLEE162 pKa = 4.22IGSKK166 pKa = 10.49FFFVDD171 pKa = 3.65KK172 pKa = 11.1SNNPVNIDD180 pKa = 4.41PISGLQSATVSKK192 pKa = 10.91LSVRR196 pKa = 11.84LSGDD200 pKa = 3.44CKK202 pKa = 10.37INQIMMSSPYY212 pKa = 9.45QIKK215 pKa = 9.64VQSEE219 pKa = 4.28EE220 pKa = 4.26PIGYY224 pKa = 9.58LIKK227 pKa = 10.67NIKK230 pKa = 9.54DD231 pKa = 3.49SKK233 pKa = 10.41LGEE236 pKa = 4.27IKK238 pKa = 9.83TVTGDD243 pKa = 3.24STINFSPDD251 pKa = 3.22EE252 pKa = 4.05LDD254 pKa = 3.76GNHH257 pKa = 6.63FLLCGDD263 pKa = 4.19RR264 pKa = 11.84SSLITKK270 pKa = 9.82VDD272 pKa = 3.19IPVRR276 pKa = 11.84NCVSKK281 pKa = 11.02FSDD284 pKa = 3.67EE285 pKa = 3.93PKK287 pKa = 10.62KK288 pKa = 10.85IFFCTNFSYY297 pKa = 10.0FKK299 pKa = 9.54WLFVFLVIFFPINWLIWKK317 pKa = 7.91TKK319 pKa = 10.1DD320 pKa = 4.25SLVVWYY326 pKa = 9.19DD327 pKa = 3.09IIGIITYY334 pKa = 9.17PILWIMNRR342 pKa = 11.84SWPYY346 pKa = 9.4FPLRR350 pKa = 11.84CRR352 pKa = 11.84ICGCFSLLTHH362 pKa = 6.65KK363 pKa = 10.75CSEE366 pKa = 4.23KK367 pKa = 10.65CVCNQSKK374 pKa = 8.94TSKK377 pKa = 8.31THH379 pKa = 4.95TSEE382 pKa = 4.37CYY384 pKa = 10.58LFIKK388 pKa = 10.54DD389 pKa = 3.42RR390 pKa = 11.84TEE392 pKa = 3.96WNCMSLIQQFQFTINTKK409 pKa = 9.58ISANFLVFITKK420 pKa = 9.52MIIASILLSYY430 pKa = 10.13LPSSMAAPKK439 pKa = 9.97SICVDD444 pKa = 2.78KK445 pKa = 11.12CYY447 pKa = 10.48FSSDD451 pKa = 3.89LKK453 pKa = 11.69SMTTSRR459 pKa = 11.84SGLSNNQYY467 pKa = 7.49EE468 pKa = 4.52TCDD471 pKa = 3.63CSIGNVITEE480 pKa = 4.58TIYY483 pKa = 10.81QEE485 pKa = 4.58GIPVSRR491 pKa = 11.84ATSVNNCVAGSTACMSEE508 pKa = 4.16GNQAEE513 pKa = 4.18NLFACRR519 pKa = 11.84YY520 pKa = 7.87GCHH523 pKa = 6.27SLVSIKK529 pKa = 10.49NIPEE533 pKa = 4.1TEE535 pKa = 3.98FVPQYY540 pKa = 10.86KK541 pKa = 10.53GISFSGNLTSLKK553 pKa = 9.82IANRR557 pKa = 11.84LRR559 pKa = 11.84NGYY562 pKa = 10.01LDD564 pKa = 4.84DD565 pKa = 4.44SSEE568 pKa = 4.28SKK570 pKa = 10.56ILEE573 pKa = 4.27KK574 pKa = 10.72EE575 pKa = 3.84SSKK578 pKa = 10.72EE579 pKa = 3.62YY580 pKa = 10.87KK581 pKa = 9.9FYY583 pKa = 9.97KK584 pKa = 8.1TLKK587 pKa = 10.57VEE589 pKa = 5.38DD590 pKa = 4.34IPPEE594 pKa = 3.93NLMPRR599 pKa = 11.84QSLVFSSEE607 pKa = 3.33VDD609 pKa = 3.03GKK611 pKa = 9.39YY612 pKa = 10.27RR613 pKa = 11.84YY614 pKa = 9.01MIEE617 pKa = 3.83MDD619 pKa = 3.39IKK621 pKa = 10.95AKK623 pKa = 9.87TGSIYY628 pKa = 10.82LLNDD632 pKa = 3.94DD633 pKa = 4.53STHH636 pKa = 7.34SPMEE640 pKa = 3.25FMIYY644 pKa = 9.46VKK646 pKa = 10.49SVGVEE651 pKa = 3.51YY652 pKa = 10.47DD653 pKa = 3.3VKK655 pKa = 11.11YY656 pKa = 10.55KK657 pKa = 11.09YY658 pKa = 9.57STAKK662 pKa = 9.93IDD664 pKa = 3.79TTISDD669 pKa = 4.15YY670 pKa = 11.45LVTCTGTCADD680 pKa = 4.67CRR682 pKa = 11.84KK683 pKa = 9.69QKK685 pKa = 10.69PKK687 pKa = 10.63VGKK690 pKa = 10.3LDD692 pKa = 3.61FCVVPTSWWGCEE704 pKa = 3.86EE705 pKa = 4.24VGCLAINEE713 pKa = 4.33GAICGHH719 pKa = 5.94CTNVYY724 pKa = 10.35DD725 pKa = 5.55LSSTVNIYY733 pKa = 9.29QVIQSHH739 pKa = 5.04VTAEE743 pKa = 3.95ICIKK747 pKa = 10.83SIDD750 pKa = 3.82GYY752 pKa = 10.32NCKK755 pKa = 9.89KK756 pKa = 10.27HH757 pKa = 6.6SDD759 pKa = 3.51RR760 pKa = 11.84SPIQTDD766 pKa = 3.95HH767 pKa = 6.6YY768 pKa = 9.96QLDD771 pKa = 3.97MTVDD775 pKa = 3.35LHH777 pKa = 7.06NDD779 pKa = 3.14YY780 pKa = 10.9MSTDD784 pKa = 3.08KK785 pKa = 11.27LFAVNKK791 pKa = 6.4QQKK794 pKa = 9.15ILTGSISDD802 pKa = 4.26LGDD805 pKa = 3.6FSSSAFGHH813 pKa = 5.32PQITIDD819 pKa = 3.86GTPLSVPSSLGQNDD833 pKa = 3.99FTWSCSAIGEE843 pKa = 4.23KK844 pKa = 10.16KK845 pKa = 10.45INIRR849 pKa = 11.84QCGLYY854 pKa = 8.99TYY856 pKa = 10.59SAIYY860 pKa = 9.95VLSTSKK866 pKa = 10.61DD867 pKa = 3.53YY868 pKa = 11.7SVMDD872 pKa = 3.65EE873 pKa = 4.57EE874 pKa = 4.97SNKK877 pKa = 10.68LYY879 pKa = 9.94MEE881 pKa = 4.67KK882 pKa = 10.79DD883 pKa = 3.46FLVGKK888 pKa = 9.62LKK890 pKa = 11.0VVVEE894 pKa = 4.34MPKK897 pKa = 10.84EE898 pKa = 4.05MFKK901 pKa = 10.89KK902 pKa = 10.52LPSKK906 pKa = 10.69PMLSEE911 pKa = 3.79TRR913 pKa = 11.84AVCSGCAQCAMGISCNLTYY932 pKa = 10.65TSDD935 pKa = 3.43TTFSARR941 pKa = 11.84LMMDD945 pKa = 3.24SCSFKK950 pKa = 10.49SDD952 pKa = 3.42QIGTVLGPNEE962 pKa = 4.27KK963 pKa = 10.32NIKK966 pKa = 9.66AYY968 pKa = 10.13CSNEE972 pKa = 3.86IQSKK976 pKa = 7.57SLKK979 pKa = 10.26LIPEE983 pKa = 4.57DD984 pKa = 3.8QSEE987 pKa = 4.53LTVDD991 pKa = 3.5IPIDD995 pKa = 3.62EE996 pKa = 4.86FVPADD1001 pKa = 3.67QDD1003 pKa = 4.22TIIHH1007 pKa = 6.6FDD1009 pKa = 3.87DD1010 pKa = 5.37KK1011 pKa = 11.3SAHH1014 pKa = 6.98DD1015 pKa = 4.17EE1016 pKa = 4.3NKK1018 pKa = 9.75HH1019 pKa = 6.53HH1020 pKa = 7.32SDD1022 pKa = 2.98TSMATLWDD1030 pKa = 3.48WVKK1033 pKa = 11.32APFNWVASFFGTFFDD1048 pKa = 3.51MVRR1051 pKa = 11.84IVLVVIAICIGIYY1064 pKa = 9.7ILSYY1068 pKa = 10.12VYY1070 pKa = 10.6KK1071 pKa = 10.77LSRR1074 pKa = 11.84SYY1076 pKa = 11.21YY1077 pKa = 8.44NEE1079 pKa = 3.55KK1080 pKa = 10.1RR1081 pKa = 11.84KK1082 pKa = 10.2HH1083 pKa = 6.48KK1084 pKa = 9.98MEE1086 pKa = 5.2DD1087 pKa = 3.37SISAIEE1093 pKa = 5.05SDD1095 pKa = 4.49LLLNDD1100 pKa = 3.58RR1101 pKa = 11.84TGMISTRR1108 pKa = 11.84KK1109 pKa = 9.65RR1110 pKa = 11.84NPPPKK1115 pKa = 9.94NYY1117 pKa = 9.73QFSLEE1122 pKa = 4.08LL1123 pKa = 4.36
MM1 pKa = 7.43KK2 pKa = 10.11KK3 pKa = 10.44YY4 pKa = 10.55YY5 pKa = 10.58LPLYY9 pKa = 9.91CLSLLLLFFVSEE21 pKa = 4.29VFLLSKK27 pKa = 9.69TEE29 pKa = 3.94HH30 pKa = 6.97DD31 pKa = 4.1SEE33 pKa = 4.03ITRR36 pKa = 11.84IRR38 pKa = 11.84DD39 pKa = 3.51RR40 pKa = 11.84YY41 pKa = 9.72PVDD44 pKa = 4.83DD45 pKa = 5.4PDD47 pKa = 5.87DD48 pKa = 3.92LVEE51 pKa = 4.94NYY53 pKa = 10.76GEE55 pKa = 4.46YY56 pKa = 9.98IPHH59 pKa = 6.53RR60 pKa = 11.84TPEE63 pKa = 3.96ADD65 pKa = 3.43KK66 pKa = 8.95PTKK69 pKa = 10.02VLSRR73 pKa = 11.84ILRR76 pKa = 11.84DD77 pKa = 3.46SSSTEE82 pKa = 3.6APRR85 pKa = 11.84LDD87 pKa = 4.97SFDD90 pKa = 3.49CMKK93 pKa = 10.53FEE95 pKa = 5.84KK96 pKa = 9.09KK97 pKa = 9.42QCMIKK102 pKa = 10.61GLSDD106 pKa = 3.94FNAHH110 pKa = 5.15YY111 pKa = 10.71QIDD114 pKa = 3.95NGNEE118 pKa = 3.73IVSCISDD125 pKa = 3.47SPNIFEE131 pKa = 3.85ICQYY135 pKa = 10.55EE136 pKa = 4.44KK137 pKa = 10.93EE138 pKa = 4.22FKK140 pKa = 9.05KK141 pKa = 9.42TKK143 pKa = 9.73FSKK146 pKa = 10.89LPVLPVLKK154 pKa = 10.71LEE156 pKa = 4.26NKK158 pKa = 9.53RR159 pKa = 11.84VLEE162 pKa = 4.22IGSKK166 pKa = 10.49FFFVDD171 pKa = 3.65KK172 pKa = 11.1SNNPVNIDD180 pKa = 4.41PISGLQSATVSKK192 pKa = 10.91LSVRR196 pKa = 11.84LSGDD200 pKa = 3.44CKK202 pKa = 10.37INQIMMSSPYY212 pKa = 9.45QIKK215 pKa = 9.64VQSEE219 pKa = 4.28EE220 pKa = 4.26PIGYY224 pKa = 9.58LIKK227 pKa = 10.67NIKK230 pKa = 9.54DD231 pKa = 3.49SKK233 pKa = 10.41LGEE236 pKa = 4.27IKK238 pKa = 9.83TVTGDD243 pKa = 3.24STINFSPDD251 pKa = 3.22EE252 pKa = 4.05LDD254 pKa = 3.76GNHH257 pKa = 6.63FLLCGDD263 pKa = 4.19RR264 pKa = 11.84SSLITKK270 pKa = 9.82VDD272 pKa = 3.19IPVRR276 pKa = 11.84NCVSKK281 pKa = 11.02FSDD284 pKa = 3.67EE285 pKa = 3.93PKK287 pKa = 10.62KK288 pKa = 10.85IFFCTNFSYY297 pKa = 10.0FKK299 pKa = 9.54WLFVFLVIFFPINWLIWKK317 pKa = 7.91TKK319 pKa = 10.1DD320 pKa = 4.25SLVVWYY326 pKa = 9.19DD327 pKa = 3.09IIGIITYY334 pKa = 9.17PILWIMNRR342 pKa = 11.84SWPYY346 pKa = 9.4FPLRR350 pKa = 11.84CRR352 pKa = 11.84ICGCFSLLTHH362 pKa = 6.65KK363 pKa = 10.75CSEE366 pKa = 4.23KK367 pKa = 10.65CVCNQSKK374 pKa = 8.94TSKK377 pKa = 8.31THH379 pKa = 4.95TSEE382 pKa = 4.37CYY384 pKa = 10.58LFIKK388 pKa = 10.54DD389 pKa = 3.42RR390 pKa = 11.84TEE392 pKa = 3.96WNCMSLIQQFQFTINTKK409 pKa = 9.58ISANFLVFITKK420 pKa = 9.52MIIASILLSYY430 pKa = 10.13LPSSMAAPKK439 pKa = 9.97SICVDD444 pKa = 2.78KK445 pKa = 11.12CYY447 pKa = 10.48FSSDD451 pKa = 3.89LKK453 pKa = 11.69SMTTSRR459 pKa = 11.84SGLSNNQYY467 pKa = 7.49EE468 pKa = 4.52TCDD471 pKa = 3.63CSIGNVITEE480 pKa = 4.58TIYY483 pKa = 10.81QEE485 pKa = 4.58GIPVSRR491 pKa = 11.84ATSVNNCVAGSTACMSEE508 pKa = 4.16GNQAEE513 pKa = 4.18NLFACRR519 pKa = 11.84YY520 pKa = 7.87GCHH523 pKa = 6.27SLVSIKK529 pKa = 10.49NIPEE533 pKa = 4.1TEE535 pKa = 3.98FVPQYY540 pKa = 10.86KK541 pKa = 10.53GISFSGNLTSLKK553 pKa = 9.82IANRR557 pKa = 11.84LRR559 pKa = 11.84NGYY562 pKa = 10.01LDD564 pKa = 4.84DD565 pKa = 4.44SSEE568 pKa = 4.28SKK570 pKa = 10.56ILEE573 pKa = 4.27KK574 pKa = 10.72EE575 pKa = 3.84SSKK578 pKa = 10.72EE579 pKa = 3.62YY580 pKa = 10.87KK581 pKa = 9.9FYY583 pKa = 9.97KK584 pKa = 8.1TLKK587 pKa = 10.57VEE589 pKa = 5.38DD590 pKa = 4.34IPPEE594 pKa = 3.93NLMPRR599 pKa = 11.84QSLVFSSEE607 pKa = 3.33VDD609 pKa = 3.03GKK611 pKa = 9.39YY612 pKa = 10.27RR613 pKa = 11.84YY614 pKa = 9.01MIEE617 pKa = 3.83MDD619 pKa = 3.39IKK621 pKa = 10.95AKK623 pKa = 9.87TGSIYY628 pKa = 10.82LLNDD632 pKa = 3.94DD633 pKa = 4.53STHH636 pKa = 7.34SPMEE640 pKa = 3.25FMIYY644 pKa = 9.46VKK646 pKa = 10.49SVGVEE651 pKa = 3.51YY652 pKa = 10.47DD653 pKa = 3.3VKK655 pKa = 11.11YY656 pKa = 10.55KK657 pKa = 11.09YY658 pKa = 9.57STAKK662 pKa = 9.93IDD664 pKa = 3.79TTISDD669 pKa = 4.15YY670 pKa = 11.45LVTCTGTCADD680 pKa = 4.67CRR682 pKa = 11.84KK683 pKa = 9.69QKK685 pKa = 10.69PKK687 pKa = 10.63VGKK690 pKa = 10.3LDD692 pKa = 3.61FCVVPTSWWGCEE704 pKa = 3.86EE705 pKa = 4.24VGCLAINEE713 pKa = 4.33GAICGHH719 pKa = 5.94CTNVYY724 pKa = 10.35DD725 pKa = 5.55LSSTVNIYY733 pKa = 9.29QVIQSHH739 pKa = 5.04VTAEE743 pKa = 3.95ICIKK747 pKa = 10.83SIDD750 pKa = 3.82GYY752 pKa = 10.32NCKK755 pKa = 9.89KK756 pKa = 10.27HH757 pKa = 6.6SDD759 pKa = 3.51RR760 pKa = 11.84SPIQTDD766 pKa = 3.95HH767 pKa = 6.6YY768 pKa = 9.96QLDD771 pKa = 3.97MTVDD775 pKa = 3.35LHH777 pKa = 7.06NDD779 pKa = 3.14YY780 pKa = 10.9MSTDD784 pKa = 3.08KK785 pKa = 11.27LFAVNKK791 pKa = 6.4QQKK794 pKa = 9.15ILTGSISDD802 pKa = 4.26LGDD805 pKa = 3.6FSSSAFGHH813 pKa = 5.32PQITIDD819 pKa = 3.86GTPLSVPSSLGQNDD833 pKa = 3.99FTWSCSAIGEE843 pKa = 4.23KK844 pKa = 10.16KK845 pKa = 10.45INIRR849 pKa = 11.84QCGLYY854 pKa = 8.99TYY856 pKa = 10.59SAIYY860 pKa = 9.95VLSTSKK866 pKa = 10.61DD867 pKa = 3.53YY868 pKa = 11.7SVMDD872 pKa = 3.65EE873 pKa = 4.57EE874 pKa = 4.97SNKK877 pKa = 10.68LYY879 pKa = 9.94MEE881 pKa = 4.67KK882 pKa = 10.79DD883 pKa = 3.46FLVGKK888 pKa = 9.62LKK890 pKa = 11.0VVVEE894 pKa = 4.34MPKK897 pKa = 10.84EE898 pKa = 4.05MFKK901 pKa = 10.89KK902 pKa = 10.52LPSKK906 pKa = 10.69PMLSEE911 pKa = 3.79TRR913 pKa = 11.84AVCSGCAQCAMGISCNLTYY932 pKa = 10.65TSDD935 pKa = 3.43TTFSARR941 pKa = 11.84LMMDD945 pKa = 3.24SCSFKK950 pKa = 10.49SDD952 pKa = 3.42QIGTVLGPNEE962 pKa = 4.27KK963 pKa = 10.32NIKK966 pKa = 9.66AYY968 pKa = 10.13CSNEE972 pKa = 3.86IQSKK976 pKa = 7.57SLKK979 pKa = 10.26LIPEE983 pKa = 4.57DD984 pKa = 3.8QSEE987 pKa = 4.53LTVDD991 pKa = 3.5IPIDD995 pKa = 3.62EE996 pKa = 4.86FVPADD1001 pKa = 3.67QDD1003 pKa = 4.22TIIHH1007 pKa = 6.6FDD1009 pKa = 3.87DD1010 pKa = 5.37KK1011 pKa = 11.3SAHH1014 pKa = 6.98DD1015 pKa = 4.17EE1016 pKa = 4.3NKK1018 pKa = 9.75HH1019 pKa = 6.53HH1020 pKa = 7.32SDD1022 pKa = 2.98TSMATLWDD1030 pKa = 3.48WVKK1033 pKa = 11.32APFNWVASFFGTFFDD1048 pKa = 3.51MVRR1051 pKa = 11.84IVLVVIAICIGIYY1064 pKa = 9.7ILSYY1068 pKa = 10.12VYY1070 pKa = 10.6KK1071 pKa = 10.77LSRR1074 pKa = 11.84SYY1076 pKa = 11.21YY1077 pKa = 8.44NEE1079 pKa = 3.55KK1080 pKa = 10.1RR1081 pKa = 11.84KK1082 pKa = 10.2HH1083 pKa = 6.48KK1084 pKa = 9.98MEE1086 pKa = 5.2DD1087 pKa = 3.37SISAIEE1093 pKa = 5.05SDD1095 pKa = 4.49LLLNDD1100 pKa = 3.58RR1101 pKa = 11.84TGMISTRR1108 pKa = 11.84KK1109 pKa = 9.65RR1110 pKa = 11.84NPPPKK1115 pKa = 9.94NYY1117 pKa = 9.73QFSLEE1122 pKa = 4.08LL1123 pKa = 4.36
Molecular weight: 127.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H4WNB2|A0A2H4WNB2_9VIRU NSm OS=Calla lily chlorotic spot virus OX=309542 PE=4 SV=1
MM1 pKa = 7.39SNVRR5 pKa = 11.84GLTHH9 pKa = 6.57QKK11 pKa = 9.93IKK13 pKa = 10.81EE14 pKa = 3.91LLAGGKK20 pKa = 9.93ADD22 pKa = 4.36VEE24 pKa = 4.4IEE26 pKa = 4.6SDD28 pKa = 3.54EE29 pKa = 4.0QTQGFNFSTFYY40 pKa = 10.78EE41 pKa = 4.59SNKK44 pKa = 10.24SGPEE48 pKa = 3.74FTFTTGINILKK59 pKa = 9.96CRR61 pKa = 11.84KK62 pKa = 9.08QVFAACKK69 pKa = 7.66SGKK72 pKa = 9.98YY73 pKa = 8.1EE74 pKa = 3.9FCGKK78 pKa = 10.13KK79 pKa = 10.07VVATTDD85 pKa = 3.42EE86 pKa = 4.36VSATDD91 pKa = 3.22WTFKK95 pKa = 9.7RR96 pKa = 11.84TEE98 pKa = 3.61AFIRR102 pKa = 11.84ARR104 pKa = 11.84LISMVEE110 pKa = 4.01HH111 pKa = 6.6TSDD114 pKa = 3.36EE115 pKa = 4.22ATKK118 pKa = 10.19KK119 pKa = 10.24QMYY122 pKa = 8.36IKK124 pKa = 10.74AMEE127 pKa = 4.47LPLVVAYY134 pKa = 9.8GLNVPAEE141 pKa = 4.32FNSAALRR148 pKa = 11.84LMLCIGGPLPLLSSIPGLAPVCFPLAYY175 pKa = 9.5FQNVKK180 pKa = 10.4KK181 pKa = 10.14EE182 pKa = 3.74QLGIKK187 pKa = 10.0NFSTYY192 pKa = 8.49EE193 pKa = 3.97QICKK197 pKa = 8.74VAKK200 pKa = 9.55VLSAASVEE208 pKa = 5.13FIDD211 pKa = 4.22KK212 pKa = 10.31TKK214 pKa = 11.15DD215 pKa = 3.39LFDD218 pKa = 3.8STVKK222 pKa = 10.91LLGDD226 pKa = 4.41SNPGTAGAISLHH238 pKa = 6.63KK239 pKa = 10.8YY240 pKa = 9.96NNQLKK245 pKa = 10.19QMEE248 pKa = 4.39TAFNSKK254 pKa = 8.66LTVDD258 pKa = 5.29DD259 pKa = 5.34FGQNSKK265 pKa = 10.14QVVKK269 pKa = 10.84KK270 pKa = 10.78KK271 pKa = 9.49NTDD274 pKa = 3.26LALL277 pKa = 4.16
MM1 pKa = 7.39SNVRR5 pKa = 11.84GLTHH9 pKa = 6.57QKK11 pKa = 9.93IKK13 pKa = 10.81EE14 pKa = 3.91LLAGGKK20 pKa = 9.93ADD22 pKa = 4.36VEE24 pKa = 4.4IEE26 pKa = 4.6SDD28 pKa = 3.54EE29 pKa = 4.0QTQGFNFSTFYY40 pKa = 10.78EE41 pKa = 4.59SNKK44 pKa = 10.24SGPEE48 pKa = 3.74FTFTTGINILKK59 pKa = 9.96CRR61 pKa = 11.84KK62 pKa = 9.08QVFAACKK69 pKa = 7.66SGKK72 pKa = 9.98YY73 pKa = 8.1EE74 pKa = 3.9FCGKK78 pKa = 10.13KK79 pKa = 10.07VVATTDD85 pKa = 3.42EE86 pKa = 4.36VSATDD91 pKa = 3.22WTFKK95 pKa = 9.7RR96 pKa = 11.84TEE98 pKa = 3.61AFIRR102 pKa = 11.84ARR104 pKa = 11.84LISMVEE110 pKa = 4.01HH111 pKa = 6.6TSDD114 pKa = 3.36EE115 pKa = 4.22ATKK118 pKa = 10.19KK119 pKa = 10.24QMYY122 pKa = 8.36IKK124 pKa = 10.74AMEE127 pKa = 4.47LPLVVAYY134 pKa = 9.8GLNVPAEE141 pKa = 4.32FNSAALRR148 pKa = 11.84LMLCIGGPLPLLSSIPGLAPVCFPLAYY175 pKa = 9.5FQNVKK180 pKa = 10.4KK181 pKa = 10.14EE182 pKa = 3.74QLGIKK187 pKa = 10.0NFSTYY192 pKa = 8.49EE193 pKa = 3.97QICKK197 pKa = 8.74VAKK200 pKa = 9.55VLSAASVEE208 pKa = 5.13FIDD211 pKa = 4.22KK212 pKa = 10.31TKK214 pKa = 11.15DD215 pKa = 3.39LFDD218 pKa = 3.8STVKK222 pKa = 10.91LLGDD226 pKa = 4.41SNPGTAGAISLHH238 pKa = 6.63KK239 pKa = 10.8YY240 pKa = 9.96NNQLKK245 pKa = 10.19QMEE248 pKa = 4.39TAFNSKK254 pKa = 8.66LTVDD258 pKa = 5.29DD259 pKa = 5.34FGQNSKK265 pKa = 10.14QVVKK269 pKa = 10.84KK270 pKa = 10.78KK271 pKa = 9.49NTDD274 pKa = 3.26LALL277 pKa = 4.16
Molecular weight: 30.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5052 |
277 |
2883 |
1010.4 |
115.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.018 ± 0.569 | 2.415 ± 0.495 |
5.8 ± 0.286 | 6.453 ± 0.488 |
4.889 ± 0.18 | 4.078 ± 0.366 |
1.92 ± 0.196 | 8.036 ± 0.378 |
8.888 ± 0.317 | 8.571 ± 0.492 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.662 ± 0.639 | 6.433 ± 0.643 |
3.088 ± 0.485 | 2.831 ± 0.2 |
3.207 ± 0.135 | 9.857 ± 0.698 |
5.562 ± 0.21 | 5.523 ± 0.363 |
0.792 ± 0.172 | 3.979 ± 0.413 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
