
Methylophilaceae phage P19250A
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
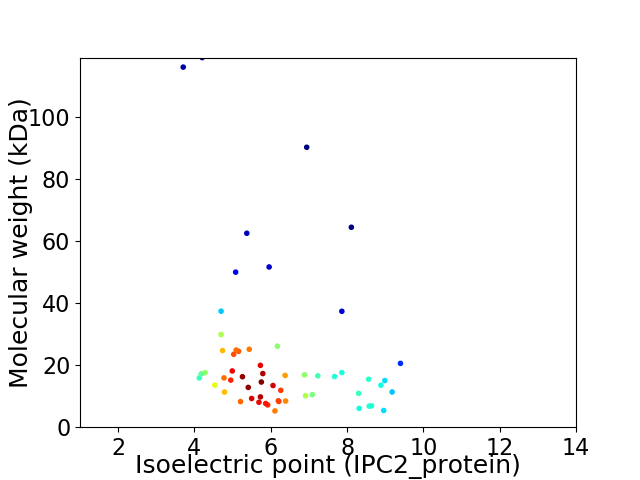
Virtual 2D-PAGE plot for 58 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H4FYM5|A0A2H4FYM5_9CAUD Uncharacterized protein OS=Methylophilaceae phage P19250A OX=1913040 GN=P19250A_0033 PE=4 SV=1
MM1 pKa = 7.29ATLSITPAPANDD13 pKa = 3.02IAVSVNATDD22 pKa = 3.51VTLSQGTTLNVEE34 pKa = 4.38VTPTPATTVVVDD46 pKa = 4.32RR47 pKa = 11.84GVTGASGLSGYY58 pKa = 10.0SGYY61 pKa = 10.67SGYY64 pKa = 10.71SGYY67 pKa = 11.05SGFSGISGASGISGFSGISGYY88 pKa = 10.66SGEE91 pKa = 4.51QGTSINIVGSVATPEE106 pKa = 4.31DD107 pKa = 4.32LPPSGNLNDD116 pKa = 4.2AYY118 pKa = 9.72IVQSNGDD125 pKa = 3.82LYY127 pKa = 11.61VWDD130 pKa = 4.37GSAWINVGQIVGPQGEE146 pKa = 4.52SGYY149 pKa = 10.59SGYY152 pKa = 11.02SGISGFSGNSGISGYY167 pKa = 10.98SGFSGVNGASGFSGEE182 pKa = 4.19SGYY185 pKa = 11.34SGFSGISGFSGEE197 pKa = 4.54SGISGWSGYY206 pKa = 10.76SGINGLSGYY215 pKa = 10.57SGFNGFSGISGWSGEE230 pKa = 4.49SGFSGYY236 pKa = 10.55SGASGISGWSGDD248 pKa = 3.51SGISGFSGFSGDD260 pKa = 3.88SGISGYY266 pKa = 10.98SGFSGISGYY275 pKa = 10.78SGASGISGWSGEE287 pKa = 4.16IGASGYY293 pKa = 10.8SGISGWSGDD302 pKa = 3.51SGISGFSGEE311 pKa = 4.66SGASGISGFSGWSGISGYY329 pKa = 10.73SGISGYY335 pKa = 10.97SGINGLSGYY344 pKa = 10.41SGINGASGEE353 pKa = 4.29SGYY356 pKa = 10.95SGWSGEE362 pKa = 3.96IGASGISGYY371 pKa = 10.44SGYY374 pKa = 11.0SGFNGAIGSSGISGFSGYY392 pKa = 10.12SGEE395 pKa = 4.49IGASGDD401 pKa = 3.57SGFSGWSGEE410 pKa = 4.12VGASGISGFSGYY422 pKa = 10.69SGFSGHH428 pKa = 6.09SGEE431 pKa = 4.68VGASGISGYY440 pKa = 10.82SGFSGEE446 pKa = 4.34VGASGDD452 pKa = 3.73SGFSGWSGEE461 pKa = 3.92IGASGISGYY470 pKa = 10.83SGISGFSGYY479 pKa = 10.11SGQIGASGFSGISGFSGFEE498 pKa = 3.95GASGHH503 pKa = 6.01SGYY506 pKa = 10.74SGWSGEE512 pKa = 4.17IGTSGHH518 pKa = 6.27SGLSGFSGFSGYY530 pKa = 10.77SGDD533 pKa = 4.08SGFSGYY539 pKa = 10.3SGEE542 pKa = 4.28SGYY545 pKa = 11.27SGAQGATGGSSSLFLYY561 pKa = 10.41KK562 pKa = 10.24ADD564 pKa = 3.61NTATSGEE571 pKa = 4.28PSPGHH576 pKa = 5.61VLWNNATQISATQINIDD593 pKa = 4.58HH594 pKa = 6.28LTEE597 pKa = 5.3DD598 pKa = 3.96NTDD601 pKa = 2.67IDD603 pKa = 3.6IFLAGLEE610 pKa = 4.09NTEE613 pKa = 3.94QFTIQDD619 pKa = 3.86RR620 pKa = 11.84NVSSNNQVWLVNGTPTNINPGTSNSYY646 pKa = 6.67WTVPVSLVSSNGTGTTGFANNQQLFLAIINGISGYY681 pKa = 10.58SGFSGYY687 pKa = 10.35SGYY690 pKa = 10.99SGAQGTSGFSGYY702 pKa = 10.28SGYY705 pKa = 11.03SGLQGNDD712 pKa = 2.9GFSGYY717 pKa = 10.11SGYY720 pKa = 10.7SGYY723 pKa = 10.85SGLVGDD729 pKa = 4.97SGYY732 pKa = 10.54SGHH735 pKa = 6.69SGISGFSGYY744 pKa = 10.11SGQVGQSGYY753 pKa = 10.26SGYY756 pKa = 10.41SGEE759 pKa = 4.22VGALGISGYY768 pKa = 10.16SGYY771 pKa = 10.54SGYY774 pKa = 10.85SGLVGDD780 pKa = 4.97SGYY783 pKa = 10.54SGHH786 pKa = 6.66SGTSGFSGWSGAVGQSGYY804 pKa = 10.63SGYY807 pKa = 10.99SGLQGNDD814 pKa = 3.11GLSGYY819 pKa = 9.79SGYY822 pKa = 10.68SGYY825 pKa = 10.85SGLVGDD831 pKa = 4.99SGYY834 pKa = 10.98SGYY837 pKa = 11.06SGFSGFSGEE846 pKa = 4.56KK847 pKa = 9.78GDD849 pKa = 4.14SGFSGFSGEE858 pKa = 4.05QGTPGLSGYY867 pKa = 9.96SGYY870 pKa = 10.98SGAQGPEE877 pKa = 3.74GHH879 pKa = 6.27SGYY882 pKa = 10.82SGINGASGISGFSGAQGEE900 pKa = 4.66SGISGYY906 pKa = 10.4SGYY909 pKa = 10.63SGEE912 pKa = 4.53QGTSGYY918 pKa = 10.79SGINGEE924 pKa = 4.46SGYY927 pKa = 10.62SGYY930 pKa = 10.79SGAVGASGISGYY942 pKa = 10.03SGWSGAVGISGYY954 pKa = 10.22SGYY957 pKa = 11.02SGIDD961 pKa = 3.32GANGSSGISGYY972 pKa = 10.41SGYY975 pKa = 11.04SGIDD979 pKa = 3.7GINGEE984 pKa = 4.58SGYY987 pKa = 10.72SGYY990 pKa = 10.92SGAEE994 pKa = 4.04GASGLSGYY1002 pKa = 10.03SGYY1005 pKa = 11.01SGAQGLSGYY1014 pKa = 10.48SGINGYY1020 pKa = 10.24SGISGFSGANGASGYY1035 pKa = 10.08SGYY1038 pKa = 10.9SGAQGISGYY1047 pKa = 10.13SGYY1050 pKa = 10.94SGATGPTVYY1059 pKa = 9.74PGAGMAVSTGSAWTTSKK1076 pKa = 9.85ATPTGVVVGDD1086 pKa = 4.13TDD1088 pKa = 4.02TQTLTNKK1095 pKa = 9.92RR1096 pKa = 11.84VTPRR1100 pKa = 11.84VLASTANSATPTLNTDD1116 pKa = 3.97LYY1118 pKa = 11.48DD1119 pKa = 3.45MMVITGQSVAITSFTTNLTGTPTNGQKK1146 pKa = 10.54LWISITGTGAIAITWGASFEE1166 pKa = 4.27ASTVALPTTTTSTNRR1181 pKa = 11.84LDD1183 pKa = 3.19IGFVYY1188 pKa = 10.58NVATSDD1194 pKa = 3.44WRR1196 pKa = 11.84CVAVAA1201 pKa = 4.46
MM1 pKa = 7.29ATLSITPAPANDD13 pKa = 3.02IAVSVNATDD22 pKa = 3.51VTLSQGTTLNVEE34 pKa = 4.38VTPTPATTVVVDD46 pKa = 4.32RR47 pKa = 11.84GVTGASGLSGYY58 pKa = 10.0SGYY61 pKa = 10.67SGYY64 pKa = 10.71SGYY67 pKa = 11.05SGFSGISGASGISGFSGISGYY88 pKa = 10.66SGEE91 pKa = 4.51QGTSINIVGSVATPEE106 pKa = 4.31DD107 pKa = 4.32LPPSGNLNDD116 pKa = 4.2AYY118 pKa = 9.72IVQSNGDD125 pKa = 3.82LYY127 pKa = 11.61VWDD130 pKa = 4.37GSAWINVGQIVGPQGEE146 pKa = 4.52SGYY149 pKa = 10.59SGYY152 pKa = 11.02SGISGFSGNSGISGYY167 pKa = 10.98SGFSGVNGASGFSGEE182 pKa = 4.19SGYY185 pKa = 11.34SGFSGISGFSGEE197 pKa = 4.54SGISGWSGYY206 pKa = 10.76SGINGLSGYY215 pKa = 10.57SGFNGFSGISGWSGEE230 pKa = 4.49SGFSGYY236 pKa = 10.55SGASGISGWSGDD248 pKa = 3.51SGISGFSGFSGDD260 pKa = 3.88SGISGYY266 pKa = 10.98SGFSGISGYY275 pKa = 10.78SGASGISGWSGEE287 pKa = 4.16IGASGYY293 pKa = 10.8SGISGWSGDD302 pKa = 3.51SGISGFSGEE311 pKa = 4.66SGASGISGFSGWSGISGYY329 pKa = 10.73SGISGYY335 pKa = 10.97SGINGLSGYY344 pKa = 10.41SGINGASGEE353 pKa = 4.29SGYY356 pKa = 10.95SGWSGEE362 pKa = 3.96IGASGISGYY371 pKa = 10.44SGYY374 pKa = 11.0SGFNGAIGSSGISGFSGYY392 pKa = 10.12SGEE395 pKa = 4.49IGASGDD401 pKa = 3.57SGFSGWSGEE410 pKa = 4.12VGASGISGFSGYY422 pKa = 10.69SGFSGHH428 pKa = 6.09SGEE431 pKa = 4.68VGASGISGYY440 pKa = 10.82SGFSGEE446 pKa = 4.34VGASGDD452 pKa = 3.73SGFSGWSGEE461 pKa = 3.92IGASGISGYY470 pKa = 10.83SGISGFSGYY479 pKa = 10.11SGQIGASGFSGISGFSGFEE498 pKa = 3.95GASGHH503 pKa = 6.01SGYY506 pKa = 10.74SGWSGEE512 pKa = 4.17IGTSGHH518 pKa = 6.27SGLSGFSGFSGYY530 pKa = 10.77SGDD533 pKa = 4.08SGFSGYY539 pKa = 10.3SGEE542 pKa = 4.28SGYY545 pKa = 11.27SGAQGATGGSSSLFLYY561 pKa = 10.41KK562 pKa = 10.24ADD564 pKa = 3.61NTATSGEE571 pKa = 4.28PSPGHH576 pKa = 5.61VLWNNATQISATQINIDD593 pKa = 4.58HH594 pKa = 6.28LTEE597 pKa = 5.3DD598 pKa = 3.96NTDD601 pKa = 2.67IDD603 pKa = 3.6IFLAGLEE610 pKa = 4.09NTEE613 pKa = 3.94QFTIQDD619 pKa = 3.86RR620 pKa = 11.84NVSSNNQVWLVNGTPTNINPGTSNSYY646 pKa = 6.67WTVPVSLVSSNGTGTTGFANNQQLFLAIINGISGYY681 pKa = 10.58SGFSGYY687 pKa = 10.35SGYY690 pKa = 10.99SGAQGTSGFSGYY702 pKa = 10.28SGYY705 pKa = 11.03SGLQGNDD712 pKa = 2.9GFSGYY717 pKa = 10.11SGYY720 pKa = 10.7SGYY723 pKa = 10.85SGLVGDD729 pKa = 4.97SGYY732 pKa = 10.54SGHH735 pKa = 6.69SGISGFSGYY744 pKa = 10.11SGQVGQSGYY753 pKa = 10.26SGYY756 pKa = 10.41SGEE759 pKa = 4.22VGALGISGYY768 pKa = 10.16SGYY771 pKa = 10.54SGYY774 pKa = 10.85SGLVGDD780 pKa = 4.97SGYY783 pKa = 10.54SGHH786 pKa = 6.66SGTSGFSGWSGAVGQSGYY804 pKa = 10.63SGYY807 pKa = 10.99SGLQGNDD814 pKa = 3.11GLSGYY819 pKa = 9.79SGYY822 pKa = 10.68SGYY825 pKa = 10.85SGLVGDD831 pKa = 4.99SGYY834 pKa = 10.98SGYY837 pKa = 11.06SGFSGFSGEE846 pKa = 4.56KK847 pKa = 9.78GDD849 pKa = 4.14SGFSGFSGEE858 pKa = 4.05QGTPGLSGYY867 pKa = 9.96SGYY870 pKa = 10.98SGAQGPEE877 pKa = 3.74GHH879 pKa = 6.27SGYY882 pKa = 10.82SGINGASGISGFSGAQGEE900 pKa = 4.66SGISGYY906 pKa = 10.4SGYY909 pKa = 10.63SGEE912 pKa = 4.53QGTSGYY918 pKa = 10.79SGINGEE924 pKa = 4.46SGYY927 pKa = 10.62SGYY930 pKa = 10.79SGAVGASGISGYY942 pKa = 10.03SGWSGAVGISGYY954 pKa = 10.22SGYY957 pKa = 11.02SGIDD961 pKa = 3.32GANGSSGISGYY972 pKa = 10.41SGYY975 pKa = 11.04SGIDD979 pKa = 3.7GINGEE984 pKa = 4.58SGYY987 pKa = 10.72SGYY990 pKa = 10.92SGAEE994 pKa = 4.04GASGLSGYY1002 pKa = 10.03SGYY1005 pKa = 11.01SGAQGLSGYY1014 pKa = 10.48SGINGYY1020 pKa = 10.24SGISGFSGANGASGYY1035 pKa = 10.08SGYY1038 pKa = 10.9SGAQGISGYY1047 pKa = 10.13SGYY1050 pKa = 10.94SGATGPTVYY1059 pKa = 9.74PGAGMAVSTGSAWTTSKK1076 pKa = 9.85ATPTGVVVGDD1086 pKa = 4.13TDD1088 pKa = 4.02TQTLTNKK1095 pKa = 9.92RR1096 pKa = 11.84VTPRR1100 pKa = 11.84VLASTANSATPTLNTDD1116 pKa = 3.97LYY1118 pKa = 11.48DD1119 pKa = 3.45MMVITGQSVAITSFTTNLTGTPTNGQKK1146 pKa = 10.54LWISITGTGAIAITWGASFEE1166 pKa = 4.27ASTVALPTTTTSTNRR1181 pKa = 11.84LDD1183 pKa = 3.19IGFVYY1188 pKa = 10.58NVATSDD1194 pKa = 3.44WRR1196 pKa = 11.84CVAVAA1201 pKa = 4.46
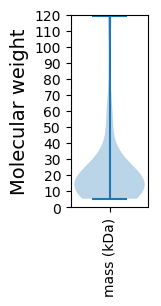
Molecular weight: 116.19 kDa
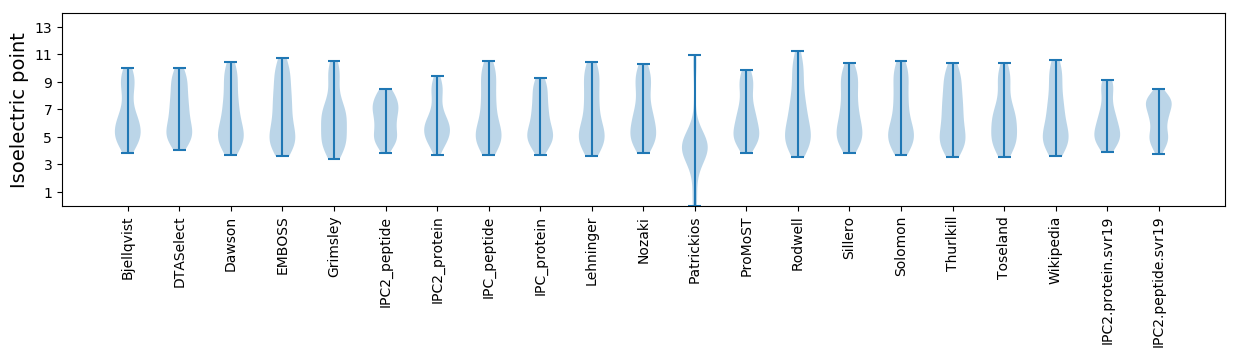
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H4FS95|A0A2H4FS95_9CAUD Uncharacterized protein OS=Methylophilaceae phage P19250A OX=1913040 GN=P19250A_0052 PE=4 SV=1
MM1 pKa = 7.21IRR3 pKa = 11.84KK4 pKa = 9.72DD5 pKa = 3.25KK6 pKa = 10.85VRR8 pKa = 11.84NTEE11 pKa = 3.34IPLYY15 pKa = 9.53YY16 pKa = 9.83WIRR19 pKa = 11.84EE20 pKa = 3.98KK21 pKa = 11.37GSINNVKK28 pKa = 9.91SGMMLGYY35 pKa = 10.2RR36 pKa = 11.84PIGWHH41 pKa = 6.1IATEE45 pKa = 4.14VLEE48 pKa = 4.67RR49 pKa = 11.84YY50 pKa = 9.33YY51 pKa = 10.74KK52 pKa = 10.87LKK54 pKa = 10.67GIKK57 pKa = 8.53PNKK60 pKa = 8.22MNKK63 pKa = 9.07FDD65 pKa = 6.32KK66 pKa = 10.34IVCDD70 pKa = 3.25AAYY73 pKa = 9.58KK74 pKa = 10.52YY75 pKa = 10.6GSKK78 pKa = 10.05PQFVEE83 pKa = 4.14ANFNFHH89 pKa = 6.13VVGDD93 pKa = 4.0RR94 pKa = 11.84EE95 pKa = 4.12LL96 pKa = 4.9
MM1 pKa = 7.21IRR3 pKa = 11.84KK4 pKa = 9.72DD5 pKa = 3.25KK6 pKa = 10.85VRR8 pKa = 11.84NTEE11 pKa = 3.34IPLYY15 pKa = 9.53YY16 pKa = 9.83WIRR19 pKa = 11.84EE20 pKa = 3.98KK21 pKa = 11.37GSINNVKK28 pKa = 9.91SGMMLGYY35 pKa = 10.2RR36 pKa = 11.84PIGWHH41 pKa = 6.1IATEE45 pKa = 4.14VLEE48 pKa = 4.67RR49 pKa = 11.84YY50 pKa = 9.33YY51 pKa = 10.74KK52 pKa = 10.87LKK54 pKa = 10.67GIKK57 pKa = 8.53PNKK60 pKa = 8.22MNKK63 pKa = 9.07FDD65 pKa = 6.32KK66 pKa = 10.34IVCDD70 pKa = 3.25AAYY73 pKa = 9.58KK74 pKa = 10.52YY75 pKa = 10.6GSKK78 pKa = 10.05PQFVEE83 pKa = 4.14ANFNFHH89 pKa = 6.13VVGDD93 pKa = 4.0RR94 pKa = 11.84EE95 pKa = 4.12LL96 pKa = 4.9
Molecular weight: 11.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12189 |
47 |
1201 |
210.2 |
23.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.892 ± 0.551 | 0.82 ± 0.127 |
5.128 ± 0.357 | 4.996 ± 0.553 |
4.291 ± 0.196 | 9.123 ± 1.885 |
1.362 ± 0.229 | 7.039 ± 0.283 |
5.53 ± 0.76 | 7.408 ± 0.511 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.322 ± 0.285 | 6.358 ± 0.412 |
3.643 ± 0.254 | 4.118 ± 0.327 |
3.191 ± 0.336 | 8.245 ± 1.388 |
7.351 ± 0.85 | 5.193 ± 0.266 |
1.493 ± 0.114 | 4.496 ± 0.365 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
