
Odonata-associated circular virus-11
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 8.16
Get precalculated fractions of proteins
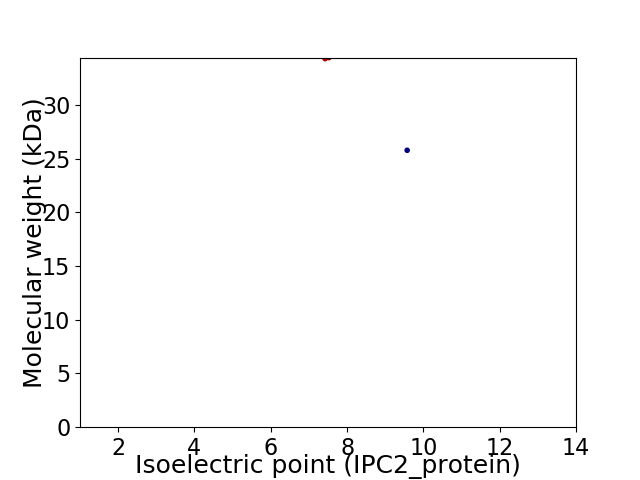
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
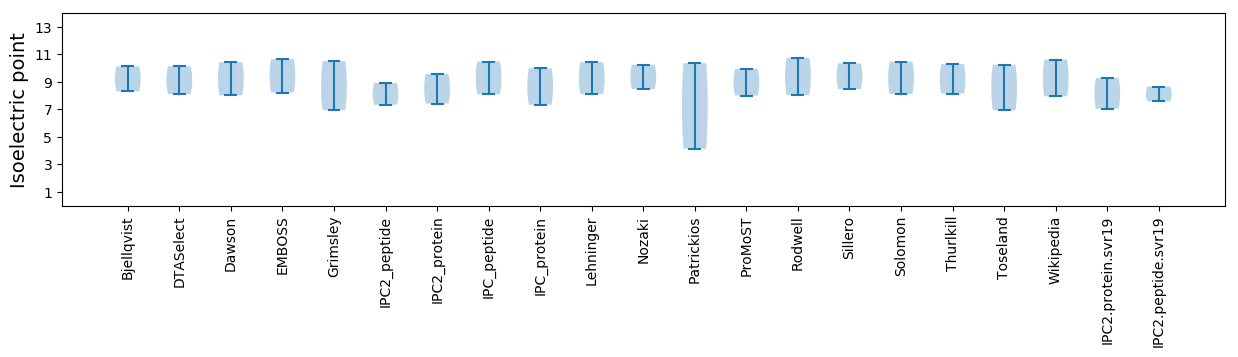
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4UH45|A0A0B4UH45_9VIRU Coat protein OS=Odonata-associated circular virus-11 OX=1592111 PE=4 SV=1
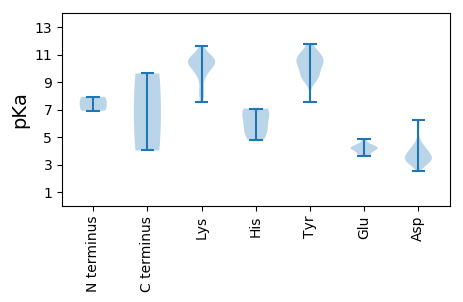
MM1 pKa = 6.92GHH3 pKa = 7.07EE4 pKa = 4.15IRR6 pKa = 11.84AKK8 pKa = 9.98SRR10 pKa = 11.84GWCFTLNNYY19 pKa = 10.41SNLEE23 pKa = 3.68ISEE26 pKa = 4.19LKK28 pKa = 10.71LRR30 pKa = 11.84FEE32 pKa = 4.18GAVWYY37 pKa = 10.02IIGEE41 pKa = 4.27EE42 pKa = 4.23VGLSGTPHH50 pKa = 6.17LQGAIYY56 pKa = 9.63YY57 pKa = 9.18KK58 pKa = 10.57NAVVMPKK65 pKa = 10.15FNPRR69 pKa = 11.84IHH71 pKa = 6.22WEE73 pKa = 3.85AMRR76 pKa = 11.84GNCQQSRR83 pKa = 11.84EE84 pKa = 4.18YY85 pKa = 10.55CSKK88 pKa = 10.62EE89 pKa = 3.81KK90 pKa = 11.11VLVEE94 pKa = 4.17YY95 pKa = 10.52GEE97 pKa = 4.46CPKK100 pKa = 10.51KK101 pKa = 10.79GKK103 pKa = 10.44RR104 pKa = 11.84SDD106 pKa = 3.24IKK108 pKa = 11.59SMVEE112 pKa = 3.59MLQAGATDD120 pKa = 3.39KK121 pKa = 10.82DD122 pKa = 3.28IYY124 pKa = 9.21MAHH127 pKa = 6.97GDD129 pKa = 3.2KK130 pKa = 10.77ALRR133 pKa = 11.84INGCLARR140 pKa = 11.84ARR142 pKa = 11.84ASLTEE147 pKa = 4.1VKK149 pKa = 10.42RR150 pKa = 11.84NWEE153 pKa = 3.61MDD155 pKa = 2.51IRR157 pKa = 11.84IYY159 pKa = 10.95YY160 pKa = 7.58GAPRR164 pKa = 11.84TGKK167 pKa = 7.55TRR169 pKa = 11.84AVYY172 pKa = 10.55EE173 pKa = 3.99EE174 pKa = 4.11FGIDD178 pKa = 3.24NVYY181 pKa = 11.28SKK183 pKa = 10.41MAGKK187 pKa = 8.48WWDD190 pKa = 3.69GYY192 pKa = 10.69RR193 pKa = 11.84GEE195 pKa = 4.39PCVLIDD201 pKa = 6.23DD202 pKa = 5.74FDD204 pKa = 4.2PQNCFDD210 pKa = 3.77IQYY213 pKa = 11.04DD214 pKa = 4.67FYY216 pKa = 11.79LKK218 pKa = 10.97LLDD221 pKa = 4.5RR222 pKa = 11.84YY223 pKa = 9.37PLRR226 pKa = 11.84IEE228 pKa = 4.12FKK230 pKa = 9.63GGSGEE235 pKa = 4.44FYY237 pKa = 10.66SKK239 pKa = 9.95TIILTSNFDD248 pKa = 3.52PAIWFTEE255 pKa = 3.91KK256 pKa = 10.82ANRR259 pKa = 11.84DD260 pKa = 3.29AFFEE264 pKa = 4.47RR265 pKa = 11.84IKK267 pKa = 10.11EE268 pKa = 3.65IRR270 pKa = 11.84YY271 pKa = 9.22FGGTEE276 pKa = 3.69VSGGNTSLLTPEE288 pKa = 4.72NKK290 pKa = 8.32PTYY293 pKa = 9.88EE294 pKa = 4.87GIGKK298 pKa = 9.62
MM1 pKa = 6.92GHH3 pKa = 7.07EE4 pKa = 4.15IRR6 pKa = 11.84AKK8 pKa = 9.98SRR10 pKa = 11.84GWCFTLNNYY19 pKa = 10.41SNLEE23 pKa = 3.68ISEE26 pKa = 4.19LKK28 pKa = 10.71LRR30 pKa = 11.84FEE32 pKa = 4.18GAVWYY37 pKa = 10.02IIGEE41 pKa = 4.27EE42 pKa = 4.23VGLSGTPHH50 pKa = 6.17LQGAIYY56 pKa = 9.63YY57 pKa = 9.18KK58 pKa = 10.57NAVVMPKK65 pKa = 10.15FNPRR69 pKa = 11.84IHH71 pKa = 6.22WEE73 pKa = 3.85AMRR76 pKa = 11.84GNCQQSRR83 pKa = 11.84EE84 pKa = 4.18YY85 pKa = 10.55CSKK88 pKa = 10.62EE89 pKa = 3.81KK90 pKa = 11.11VLVEE94 pKa = 4.17YY95 pKa = 10.52GEE97 pKa = 4.46CPKK100 pKa = 10.51KK101 pKa = 10.79GKK103 pKa = 10.44RR104 pKa = 11.84SDD106 pKa = 3.24IKK108 pKa = 11.59SMVEE112 pKa = 3.59MLQAGATDD120 pKa = 3.39KK121 pKa = 10.82DD122 pKa = 3.28IYY124 pKa = 9.21MAHH127 pKa = 6.97GDD129 pKa = 3.2KK130 pKa = 10.77ALRR133 pKa = 11.84INGCLARR140 pKa = 11.84ARR142 pKa = 11.84ASLTEE147 pKa = 4.1VKK149 pKa = 10.42RR150 pKa = 11.84NWEE153 pKa = 3.61MDD155 pKa = 2.51IRR157 pKa = 11.84IYY159 pKa = 10.95YY160 pKa = 7.58GAPRR164 pKa = 11.84TGKK167 pKa = 7.55TRR169 pKa = 11.84AVYY172 pKa = 10.55EE173 pKa = 3.99EE174 pKa = 4.11FGIDD178 pKa = 3.24NVYY181 pKa = 11.28SKK183 pKa = 10.41MAGKK187 pKa = 8.48WWDD190 pKa = 3.69GYY192 pKa = 10.69RR193 pKa = 11.84GEE195 pKa = 4.39PCVLIDD201 pKa = 6.23DD202 pKa = 5.74FDD204 pKa = 4.2PQNCFDD210 pKa = 3.77IQYY213 pKa = 11.04DD214 pKa = 4.67FYY216 pKa = 11.79LKK218 pKa = 10.97LLDD221 pKa = 4.5RR222 pKa = 11.84YY223 pKa = 9.37PLRR226 pKa = 11.84IEE228 pKa = 4.12FKK230 pKa = 9.63GGSGEE235 pKa = 4.44FYY237 pKa = 10.66SKK239 pKa = 9.95TIILTSNFDD248 pKa = 3.52PAIWFTEE255 pKa = 3.91KK256 pKa = 10.82ANRR259 pKa = 11.84DD260 pKa = 3.29AFFEE264 pKa = 4.47RR265 pKa = 11.84IKK267 pKa = 10.11EE268 pKa = 3.65IRR270 pKa = 11.84YY271 pKa = 9.22FGGTEE276 pKa = 3.69VSGGNTSLLTPEE288 pKa = 4.72NKK290 pKa = 8.32PTYY293 pKa = 9.88EE294 pKa = 4.87GIGKK298 pKa = 9.62
Molecular weight: 34.31 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UH45|A0A0B4UH45_9VIRU Coat protein OS=Odonata-associated circular virus-11 OX=1592111 PE=4 SV=1
MM1 pKa = 7.93PYY3 pKa = 10.35NRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84TYY9 pKa = 9.77RR10 pKa = 11.84RR11 pKa = 11.84KK12 pKa = 9.28RR13 pKa = 11.84NYY15 pKa = 9.38RR16 pKa = 11.84RR17 pKa = 11.84KK18 pKa = 7.85TSRR21 pKa = 11.84ASIYY25 pKa = 10.63GRR27 pKa = 11.84AGRR30 pKa = 11.84QLWKK34 pKa = 10.47DD35 pKa = 3.33VKK37 pKa = 9.46TLKK40 pKa = 10.8NLINVEE46 pKa = 4.17WKK48 pKa = 9.55VQNHH52 pKa = 4.82VTTSTSVPNTGTVMLLNHH70 pKa = 7.02LALGEE75 pKa = 4.37DD76 pKa = 3.92YY77 pKa = 11.2NNRR80 pKa = 11.84SGRR83 pKa = 11.84QVRR86 pKa = 11.84WKK88 pKa = 10.22SLQWRR93 pKa = 11.84CTTQKK98 pKa = 10.26HH99 pKa = 5.41ASATSSRR106 pKa = 11.84VRR108 pKa = 11.84MLIVIDD114 pKa = 4.27RR115 pKa = 11.84QCKK118 pKa = 8.36QALPSASEE126 pKa = 3.96ILDD129 pKa = 3.5SSVAPLIDD137 pKa = 4.01GFRR140 pKa = 11.84QLDD143 pKa = 3.23ARR145 pKa = 11.84KK146 pKa = 9.82RR147 pKa = 11.84FVILKK152 pKa = 10.43DD153 pKa = 3.83VMFLLNNDD161 pKa = 3.48RR162 pKa = 11.84PEE164 pKa = 4.11LTKK167 pKa = 10.39TGYY170 pKa = 10.82FKK172 pKa = 10.58MDD174 pKa = 3.15MKK176 pKa = 10.35TVYY179 pKa = 10.14DD180 pKa = 3.95ASDD183 pKa = 3.36NGDD186 pKa = 3.06ITDD189 pKa = 3.91IYY191 pKa = 11.15SNSLYY196 pKa = 10.2MVMIGDD202 pKa = 4.0EE203 pKa = 4.31ATNQPTHH210 pKa = 5.29EE211 pKa = 4.4TNVRR215 pKa = 11.84LRR217 pKa = 11.84YY218 pKa = 9.35IDD220 pKa = 3.37NN221 pKa = 4.02
MM1 pKa = 7.93PYY3 pKa = 10.35NRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84TYY9 pKa = 9.77RR10 pKa = 11.84RR11 pKa = 11.84KK12 pKa = 9.28RR13 pKa = 11.84NYY15 pKa = 9.38RR16 pKa = 11.84RR17 pKa = 11.84KK18 pKa = 7.85TSRR21 pKa = 11.84ASIYY25 pKa = 10.63GRR27 pKa = 11.84AGRR30 pKa = 11.84QLWKK34 pKa = 10.47DD35 pKa = 3.33VKK37 pKa = 9.46TLKK40 pKa = 10.8NLINVEE46 pKa = 4.17WKK48 pKa = 9.55VQNHH52 pKa = 4.82VTTSTSVPNTGTVMLLNHH70 pKa = 7.02LALGEE75 pKa = 4.37DD76 pKa = 3.92YY77 pKa = 11.2NNRR80 pKa = 11.84SGRR83 pKa = 11.84QVRR86 pKa = 11.84WKK88 pKa = 10.22SLQWRR93 pKa = 11.84CTTQKK98 pKa = 10.26HH99 pKa = 5.41ASATSSRR106 pKa = 11.84VRR108 pKa = 11.84MLIVIDD114 pKa = 4.27RR115 pKa = 11.84QCKK118 pKa = 8.36QALPSASEE126 pKa = 3.96ILDD129 pKa = 3.5SSVAPLIDD137 pKa = 4.01GFRR140 pKa = 11.84QLDD143 pKa = 3.23ARR145 pKa = 11.84KK146 pKa = 9.82RR147 pKa = 11.84FVILKK152 pKa = 10.43DD153 pKa = 3.83VMFLLNNDD161 pKa = 3.48RR162 pKa = 11.84PEE164 pKa = 4.11LTKK167 pKa = 10.39TGYY170 pKa = 10.82FKK172 pKa = 10.58MDD174 pKa = 3.15MKK176 pKa = 10.35TVYY179 pKa = 10.14DD180 pKa = 3.95ASDD183 pKa = 3.36NGDD186 pKa = 3.06ITDD189 pKa = 3.91IYY191 pKa = 11.15SNSLYY196 pKa = 10.2MVMIGDD202 pKa = 4.0EE203 pKa = 4.31ATNQPTHH210 pKa = 5.29EE211 pKa = 4.4TNVRR215 pKa = 11.84LRR217 pKa = 11.84YY218 pKa = 9.35IDD220 pKa = 3.37NN221 pKa = 4.02
Molecular weight: 25.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
519 |
221 |
298 |
259.5 |
30.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.588 ± 0.348 | 1.734 ± 0.473 |
5.78 ± 0.575 | 5.973 ± 1.859 |
3.468 ± 0.946 | 7.129 ± 1.744 |
1.541 ± 0.153 | 6.166 ± 0.678 |
7.129 ± 0.453 | 7.322 ± 0.728 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.083 ± 0.306 | 5.78 ± 0.833 |
3.276 ± 0.32 | 2.89 ± 0.675 |
8.478 ± 1.359 | 5.973 ± 0.723 |
5.973 ± 1.239 | 5.202 ± 0.904 |
2.119 ± 0.177 | 5.395 ± 0.497 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
