
Chthoniobacter flavus Ellin428
Taxonomy: cellular organisms; Bacteria; PVC group; Verrucomicrobia; Spartobacteria; Chthoniobacterales; Chthoniobacteraceae; Chthoniobacter; Chthoniobacter flavus
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
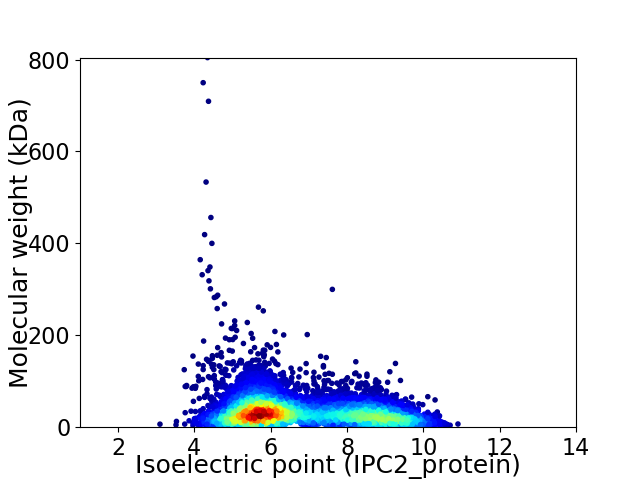
Virtual 2D-PAGE plot for 6705 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
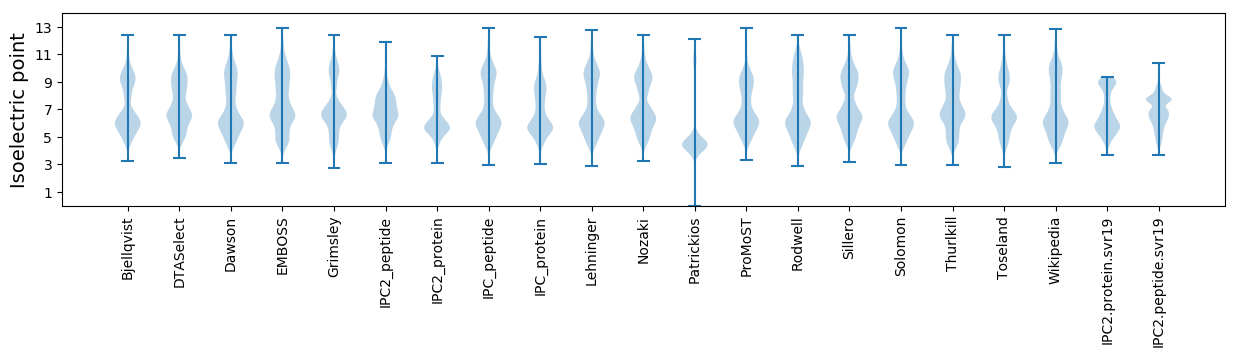
Protein with the lowest isoelectric point:
>tr|B4DBE6|B4DBE6_9BACT Uncharacterized protein OS=Chthoniobacter flavus Ellin428 OX=497964 GN=CfE428DRAFT_6237 PE=4 SV=1
MM1 pKa = 8.11DD2 pKa = 4.7EE3 pKa = 4.87SFLPTGSGTGPAGSTVDD20 pKa = 3.48TEE22 pKa = 4.36AFAGAFTHH30 pKa = 6.91VNGADD35 pKa = 3.75GASVSYY41 pKa = 10.07GLSVSASGVDD51 pKa = 3.09SGLIDD56 pKa = 3.96SLTGNHH62 pKa = 5.49VFLFLEE68 pKa = 4.38SGAVVARR75 pKa = 11.84EE76 pKa = 4.32GTTSATATTGPVEE89 pKa = 4.27FTLSVDD95 pKa = 3.11GSGNVTATLDD105 pKa = 3.59RR106 pKa = 11.84SVHH109 pKa = 5.59QGSADD114 pKa = 3.64SPVDD118 pKa = 3.73SNEE121 pKa = 3.82AVSLTSGLVTLTATITDD138 pKa = 3.54ADD140 pKa = 4.29GDD142 pKa = 4.14HH143 pKa = 6.14QAASIDD149 pKa = 3.96LGSHH153 pKa = 5.85VSIHH157 pKa = 7.5DD158 pKa = 4.61DD159 pKa = 3.91GPSITAVGTATALNVDD175 pKa = 4.16EE176 pKa = 5.67SFLPTGSGTGPTGSTVDD193 pKa = 3.49TEE195 pKa = 4.39AFAGAFTHH203 pKa = 6.91VNGADD208 pKa = 3.67GASISYY214 pKa = 9.46GLSVSASGVDD224 pKa = 3.09SGLIDD229 pKa = 3.96SLTGNHH235 pKa = 5.49VFLFLEE241 pKa = 4.38SGAVVARR248 pKa = 11.84EE249 pKa = 4.32GTTSATAATGPIEE262 pKa = 4.24FTLSVDD268 pKa = 3.12GSGNVTATLDD278 pKa = 3.66RR279 pKa = 11.84SVHH282 pKa = 5.15QSSADD287 pKa = 3.56SPVDD291 pKa = 3.68SNEE294 pKa = 3.82AVSLTSGLVTLTATITDD311 pKa = 3.54ADD313 pKa = 4.29GDD315 pKa = 4.14HH316 pKa = 6.14QAASIDD322 pKa = 3.96LGSHH326 pKa = 5.85VSIHH330 pKa = 7.5DD331 pKa = 4.61DD332 pKa = 3.91GPSITAVGTATALNVDD348 pKa = 4.16EE349 pKa = 5.67SFLPTGSGTGPAGSTVDD366 pKa = 3.48TEE368 pKa = 4.36AFAGAFTHH376 pKa = 7.28LNGADD381 pKa = 3.77GASVSYY387 pKa = 10.07GLSVSASGVDD397 pKa = 3.28SGLVDD402 pKa = 3.84SLTGNHH408 pKa = 5.48VFLFLEE414 pKa = 4.38SGAVVARR421 pKa = 11.84EE422 pKa = 4.14GTTSVTAATGPVEE435 pKa = 4.26FTLSVDD441 pKa = 3.11GSGNVTATLDD451 pKa = 3.66RR452 pKa = 11.84SVHH455 pKa = 5.15QSSADD460 pKa = 3.56SPVDD464 pKa = 3.68SNEE467 pKa = 3.82AVSLTSGLVALTATITDD484 pKa = 3.48ADD486 pKa = 4.29GDD488 pKa = 4.14HH489 pKa = 6.14QAASIDD495 pKa = 3.96LGSHH499 pKa = 5.85VSIHH503 pKa = 7.5DD504 pKa = 4.61DD505 pKa = 3.91GPSITAVGTATALNVDD521 pKa = 4.16EE522 pKa = 5.67SFLPTGSGTGPTGSTVDD539 pKa = 3.49TEE541 pKa = 4.39AFAGAFTHH549 pKa = 6.91VNGADD554 pKa = 3.67GASISYY560 pKa = 9.46GLSVSASGVDD570 pKa = 3.09SGLIDD575 pKa = 3.96SLTGNHH581 pKa = 5.49VFLFLEE587 pKa = 4.38SGAVVARR594 pKa = 11.84EE595 pKa = 4.06GATSVAAATGSIEE608 pKa = 4.07FTLSVDD614 pKa = 3.12GSGNVTATLDD624 pKa = 3.53RR625 pKa = 11.84SVHH628 pKa = 5.22QNSPDD633 pKa = 3.59SQVDD637 pKa = 3.27ANEE640 pKa = 4.2AVSLTSGLVTLTATITDD657 pKa = 3.39GDD659 pKa = 4.26GDD661 pKa = 4.05HH662 pKa = 6.33QAASIDD668 pKa = 3.96LGSHH672 pKa = 5.85VSIHH676 pKa = 7.48DD677 pKa = 4.57DD678 pKa = 3.77GPTITAIQNLIMPNIDD694 pKa = 3.09NTVDD698 pKa = 2.84HH699 pKa = 6.32GTWVPVFGADD709 pKa = 3.82GPSATSAVSVLMGTPPAGTTYY730 pKa = 10.58TVTDD734 pKa = 3.49TLTNVSGHH742 pKa = 5.9EE743 pKa = 3.98LFEE746 pKa = 4.61VDD748 pKa = 3.9VSSSTLSYY756 pKa = 10.57TFYY759 pKa = 10.71EE760 pKa = 4.17YY761 pKa = 11.17SDD763 pKa = 4.06YY764 pKa = 10.99NTLTNNGEE772 pKa = 4.05MFAFTDD778 pKa = 4.16LAHH781 pKa = 5.62TTSFFEE787 pKa = 4.65LLANTGGTYY796 pKa = 10.44SFHH799 pKa = 7.74LDD801 pKa = 3.39TNSLQSSTTISLTGGAHH818 pKa = 6.56SGTGDD823 pKa = 3.26FLTFVGTTSTFANGNDD839 pKa = 3.4PTTGYY844 pKa = 10.87DD845 pKa = 3.16ILVDD849 pKa = 3.89GWNTADD855 pKa = 4.07TNPNDD860 pKa = 3.62HH861 pKa = 6.19TVHH864 pKa = 5.81VNNNGGGIDD873 pKa = 3.71SAISIPTSPRR883 pKa = 11.84CSGSRR888 pKa = 11.84ITT890 pKa = 5.26
MM1 pKa = 8.11DD2 pKa = 4.7EE3 pKa = 4.87SFLPTGSGTGPAGSTVDD20 pKa = 3.48TEE22 pKa = 4.36AFAGAFTHH30 pKa = 6.91VNGADD35 pKa = 3.75GASVSYY41 pKa = 10.07GLSVSASGVDD51 pKa = 3.09SGLIDD56 pKa = 3.96SLTGNHH62 pKa = 5.49VFLFLEE68 pKa = 4.38SGAVVARR75 pKa = 11.84EE76 pKa = 4.32GTTSATATTGPVEE89 pKa = 4.27FTLSVDD95 pKa = 3.11GSGNVTATLDD105 pKa = 3.59RR106 pKa = 11.84SVHH109 pKa = 5.59QGSADD114 pKa = 3.64SPVDD118 pKa = 3.73SNEE121 pKa = 3.82AVSLTSGLVTLTATITDD138 pKa = 3.54ADD140 pKa = 4.29GDD142 pKa = 4.14HH143 pKa = 6.14QAASIDD149 pKa = 3.96LGSHH153 pKa = 5.85VSIHH157 pKa = 7.5DD158 pKa = 4.61DD159 pKa = 3.91GPSITAVGTATALNVDD175 pKa = 4.16EE176 pKa = 5.67SFLPTGSGTGPTGSTVDD193 pKa = 3.49TEE195 pKa = 4.39AFAGAFTHH203 pKa = 6.91VNGADD208 pKa = 3.67GASISYY214 pKa = 9.46GLSVSASGVDD224 pKa = 3.09SGLIDD229 pKa = 3.96SLTGNHH235 pKa = 5.49VFLFLEE241 pKa = 4.38SGAVVARR248 pKa = 11.84EE249 pKa = 4.32GTTSATAATGPIEE262 pKa = 4.24FTLSVDD268 pKa = 3.12GSGNVTATLDD278 pKa = 3.66RR279 pKa = 11.84SVHH282 pKa = 5.15QSSADD287 pKa = 3.56SPVDD291 pKa = 3.68SNEE294 pKa = 3.82AVSLTSGLVTLTATITDD311 pKa = 3.54ADD313 pKa = 4.29GDD315 pKa = 4.14HH316 pKa = 6.14QAASIDD322 pKa = 3.96LGSHH326 pKa = 5.85VSIHH330 pKa = 7.5DD331 pKa = 4.61DD332 pKa = 3.91GPSITAVGTATALNVDD348 pKa = 4.16EE349 pKa = 5.67SFLPTGSGTGPAGSTVDD366 pKa = 3.48TEE368 pKa = 4.36AFAGAFTHH376 pKa = 7.28LNGADD381 pKa = 3.77GASVSYY387 pKa = 10.07GLSVSASGVDD397 pKa = 3.28SGLVDD402 pKa = 3.84SLTGNHH408 pKa = 5.48VFLFLEE414 pKa = 4.38SGAVVARR421 pKa = 11.84EE422 pKa = 4.14GTTSVTAATGPVEE435 pKa = 4.26FTLSVDD441 pKa = 3.11GSGNVTATLDD451 pKa = 3.66RR452 pKa = 11.84SVHH455 pKa = 5.15QSSADD460 pKa = 3.56SPVDD464 pKa = 3.68SNEE467 pKa = 3.82AVSLTSGLVALTATITDD484 pKa = 3.48ADD486 pKa = 4.29GDD488 pKa = 4.14HH489 pKa = 6.14QAASIDD495 pKa = 3.96LGSHH499 pKa = 5.85VSIHH503 pKa = 7.5DD504 pKa = 4.61DD505 pKa = 3.91GPSITAVGTATALNVDD521 pKa = 4.16EE522 pKa = 5.67SFLPTGSGTGPTGSTVDD539 pKa = 3.49TEE541 pKa = 4.39AFAGAFTHH549 pKa = 6.91VNGADD554 pKa = 3.67GASISYY560 pKa = 9.46GLSVSASGVDD570 pKa = 3.09SGLIDD575 pKa = 3.96SLTGNHH581 pKa = 5.49VFLFLEE587 pKa = 4.38SGAVVARR594 pKa = 11.84EE595 pKa = 4.06GATSVAAATGSIEE608 pKa = 4.07FTLSVDD614 pKa = 3.12GSGNVTATLDD624 pKa = 3.53RR625 pKa = 11.84SVHH628 pKa = 5.22QNSPDD633 pKa = 3.59SQVDD637 pKa = 3.27ANEE640 pKa = 4.2AVSLTSGLVTLTATITDD657 pKa = 3.39GDD659 pKa = 4.26GDD661 pKa = 4.05HH662 pKa = 6.33QAASIDD668 pKa = 3.96LGSHH672 pKa = 5.85VSIHH676 pKa = 7.48DD677 pKa = 4.57DD678 pKa = 3.77GPTITAIQNLIMPNIDD694 pKa = 3.09NTVDD698 pKa = 2.84HH699 pKa = 6.32GTWVPVFGADD709 pKa = 3.82GPSATSAVSVLMGTPPAGTTYY730 pKa = 10.58TVTDD734 pKa = 3.49TLTNVSGHH742 pKa = 5.9EE743 pKa = 3.98LFEE746 pKa = 4.61VDD748 pKa = 3.9VSSSTLSYY756 pKa = 10.57TFYY759 pKa = 10.71EE760 pKa = 4.17YY761 pKa = 11.17SDD763 pKa = 4.06YY764 pKa = 10.99NTLTNNGEE772 pKa = 4.05MFAFTDD778 pKa = 4.16LAHH781 pKa = 5.62TTSFFEE787 pKa = 4.65LLANTGGTYY796 pKa = 10.44SFHH799 pKa = 7.74LDD801 pKa = 3.39TNSLQSSTTISLTGGAHH818 pKa = 6.56SGTGDD823 pKa = 3.26FLTFVGTTSTFANGNDD839 pKa = 3.4PTTGYY844 pKa = 10.87DD845 pKa = 3.16ILVDD849 pKa = 3.89GWNTADD855 pKa = 4.07TNPNDD860 pKa = 3.62HH861 pKa = 6.19TVHH864 pKa = 5.81VNNNGGGIDD873 pKa = 3.71SAISIPTSPRR883 pKa = 11.84CSGSRR888 pKa = 11.84ITT890 pKa = 5.26
Molecular weight: 88.65 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B4DCJ5|B4DCJ5_9BACT Tetratricopeptide TPR_4 protein OS=Chthoniobacter flavus Ellin428 OX=497964 GN=CfE428DRAFT_6636 PE=4 SV=1
MM1 pKa = 7.46KK2 pKa = 9.96RR3 pKa = 11.84QLVLRR8 pKa = 11.84VAGRR12 pKa = 11.84AASFLFFIIGTIIAQALRR30 pKa = 11.84VSFSKK35 pKa = 10.69AARR38 pKa = 11.84QRR40 pKa = 11.84RR41 pKa = 11.84ARR43 pKa = 11.84AEE45 pKa = 3.74WLQRR49 pKa = 11.84TAQRR53 pKa = 11.84CVQILRR59 pKa = 11.84LKK61 pKa = 10.21INRR64 pKa = 11.84FGALPTSGLVVANHH78 pKa = 6.86LSYY81 pKa = 10.83IDD83 pKa = 4.41IILLAAQRR91 pKa = 11.84PCVFVSKK98 pKa = 10.82SEE100 pKa = 3.95VHH102 pKa = 5.56SWPIFGQCARR112 pKa = 11.84LGGTIFVDD120 pKa = 4.05RR121 pKa = 11.84KK122 pKa = 10.31HH123 pKa = 6.8RR124 pKa = 11.84GDD126 pKa = 3.38VAGVTDD132 pKa = 4.99LMGAALDD139 pKa = 3.87EE140 pKa = 4.64GALVILFPEE149 pKa = 5.22GTSSGGASVLPFKK162 pKa = 11.09SSLLEE167 pKa = 3.92PALHH171 pKa = 6.21VNHH174 pKa = 7.32PVTAIAIAYY183 pKa = 9.13ALEE186 pKa = 4.19QGSVANEE193 pKa = 3.18ICYY196 pKa = 9.15WRR198 pKa = 11.84DD199 pKa = 3.09MTLLPHH205 pKa = 7.39LLNVWSKK212 pKa = 11.0SLIYY216 pKa = 10.43CSLRR220 pKa = 11.84CGVGRR225 pKa = 11.84QRR227 pKa = 11.84RR228 pKa = 11.84GDD230 pKa = 3.52RR231 pKa = 11.84KK232 pKa = 9.25SLARR236 pKa = 11.84EE237 pKa = 3.78LHH239 pKa = 6.59DD240 pKa = 4.27EE241 pKa = 4.21VAALHH246 pKa = 5.89AASTRR251 pKa = 11.84EE252 pKa = 3.73LAVRR256 pKa = 11.84IPQTFEE262 pKa = 3.42QPRR265 pKa = 11.84PVLMGGLPSAAKK277 pKa = 9.08PQRR280 pKa = 11.84PP281 pKa = 3.4
MM1 pKa = 7.46KK2 pKa = 9.96RR3 pKa = 11.84QLVLRR8 pKa = 11.84VAGRR12 pKa = 11.84AASFLFFIIGTIIAQALRR30 pKa = 11.84VSFSKK35 pKa = 10.69AARR38 pKa = 11.84QRR40 pKa = 11.84RR41 pKa = 11.84ARR43 pKa = 11.84AEE45 pKa = 3.74WLQRR49 pKa = 11.84TAQRR53 pKa = 11.84CVQILRR59 pKa = 11.84LKK61 pKa = 10.21INRR64 pKa = 11.84FGALPTSGLVVANHH78 pKa = 6.86LSYY81 pKa = 10.83IDD83 pKa = 4.41IILLAAQRR91 pKa = 11.84PCVFVSKK98 pKa = 10.82SEE100 pKa = 3.95VHH102 pKa = 5.56SWPIFGQCARR112 pKa = 11.84LGGTIFVDD120 pKa = 4.05RR121 pKa = 11.84KK122 pKa = 10.31HH123 pKa = 6.8RR124 pKa = 11.84GDD126 pKa = 3.38VAGVTDD132 pKa = 4.99LMGAALDD139 pKa = 3.87EE140 pKa = 4.64GALVILFPEE149 pKa = 5.22GTSSGGASVLPFKK162 pKa = 11.09SSLLEE167 pKa = 3.92PALHH171 pKa = 6.21VNHH174 pKa = 7.32PVTAIAIAYY183 pKa = 9.13ALEE186 pKa = 4.19QGSVANEE193 pKa = 3.18ICYY196 pKa = 9.15WRR198 pKa = 11.84DD199 pKa = 3.09MTLLPHH205 pKa = 7.39LLNVWSKK212 pKa = 11.0SLIYY216 pKa = 10.43CSLRR220 pKa = 11.84CGVGRR225 pKa = 11.84QRR227 pKa = 11.84RR228 pKa = 11.84GDD230 pKa = 3.52RR231 pKa = 11.84KK232 pKa = 9.25SLARR236 pKa = 11.84EE237 pKa = 3.78LHH239 pKa = 6.59DD240 pKa = 4.27EE241 pKa = 4.21VAALHH246 pKa = 5.89AASTRR251 pKa = 11.84EE252 pKa = 3.73LAVRR256 pKa = 11.84IPQTFEE262 pKa = 3.42QPRR265 pKa = 11.84PVLMGGLPSAAKK277 pKa = 9.08PQRR280 pKa = 11.84PP281 pKa = 3.4
Molecular weight: 30.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2302626 |
30 |
8197 |
343.4 |
37.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.809 ± 0.035 | 0.975 ± 0.012 |
5.223 ± 0.025 | 5.675 ± 0.05 |
4.03 ± 0.019 | 8.196 ± 0.066 |
2.373 ± 0.022 | 4.858 ± 0.024 |
4.069 ± 0.04 | 9.893 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.0 ± 0.016 | 3.386 ± 0.056 |
5.595 ± 0.04 | 3.594 ± 0.019 |
6.333 ± 0.054 | 5.87 ± 0.044 |
6.173 ± 0.077 | 6.908 ± 0.025 |
1.575 ± 0.016 | 2.466 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
