
Mushroom bacilliform virus (isolate Australia/AUS LF-1) (MBV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Barnaviridae; Barnavirus; Mushroom bacilliform virus
Average proteome isoelectric point is 7.34
Get precalculated fractions of proteins
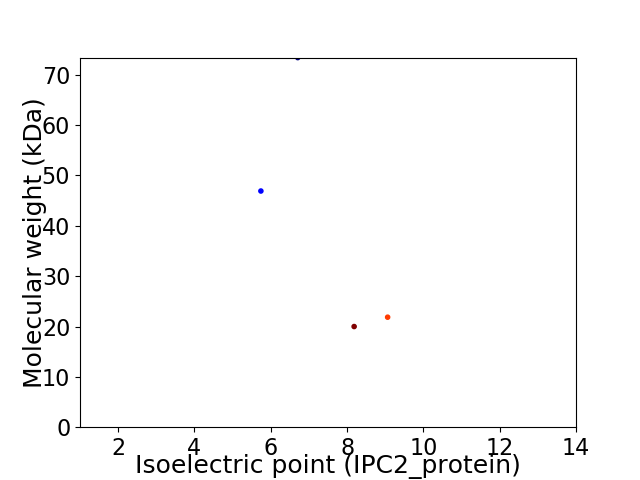
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q9YPD6|PTASE_MBVLF Putative serine protease OS=Mushroom bacilliform virus (isolate Australia/AUS LF-1) OX=650482 GN=ORF2 PE=3 SV=1
MM1 pKa = 8.48DD2 pKa = 4.88SGLYY6 pKa = 9.61PKK8 pKa = 9.96TGAPTWFYY16 pKa = 10.36GWSKK20 pKa = 9.92QRR22 pKa = 11.84EE23 pKa = 4.4GGFSDD28 pKa = 4.43FGSEE32 pKa = 3.88IFRR35 pKa = 11.84LRR37 pKa = 11.84ALPPLKK43 pKa = 10.79AEE45 pKa = 3.94IFRR48 pKa = 11.84GIRR51 pKa = 11.84EE52 pKa = 4.03VVEE55 pKa = 3.84YY56 pKa = 10.82NITGDD61 pKa = 3.62SHH63 pKa = 7.4PGYY66 pKa = 8.99PWCKK70 pKa = 9.89LGSDD74 pKa = 3.67NKK76 pKa = 10.93AVLTGFGDD84 pKa = 4.55LIWDD88 pKa = 4.0EE89 pKa = 4.01VAKK92 pKa = 10.62RR93 pKa = 11.84FNNMLGYY100 pKa = 10.8GDD102 pKa = 5.99AIFSMTPSEE111 pKa = 4.05LVQNGICDD119 pKa = 3.58AVKK122 pKa = 10.72VFIKK126 pKa = 10.35QEE128 pKa = 3.77PHH130 pKa = 6.4SLEE133 pKa = 3.93KK134 pKa = 11.1VNAGRR139 pKa = 11.84LRR141 pKa = 11.84IIAAVGLVDD150 pKa = 4.84QIVTRR155 pKa = 11.84LLCMKK160 pKa = 10.14QNNAEE165 pKa = 3.86IDD167 pKa = 3.68CWEE170 pKa = 4.5SCPSAPGMGLNDD182 pKa = 3.31EE183 pKa = 4.73GLRR186 pKa = 11.84TLYY189 pKa = 9.67STAQVMAEE197 pKa = 4.36HH198 pKa = 6.23GTICEE203 pKa = 4.16TDD205 pKa = 2.74ISGWDD210 pKa = 3.21WSVQQWEE217 pKa = 4.41LDD219 pKa = 3.01SDD221 pKa = 3.66ARR223 pKa = 11.84LRR225 pKa = 11.84TQLAGEE231 pKa = 4.39EE232 pKa = 3.81IGGYY236 pKa = 10.65LNFFLRR242 pKa = 11.84VHH244 pKa = 6.88AYY246 pKa = 9.62VVGHH250 pKa = 5.83SVFVMPDD257 pKa = 3.08GEE259 pKa = 4.37MLEE262 pKa = 4.14QTVPGGQLSGDD273 pKa = 4.01YY274 pKa = 8.36NTSSSNSRR282 pKa = 11.84MRR284 pKa = 11.84VIATMFARR292 pKa = 11.84YY293 pKa = 8.52LAGQVSGFPLLGIKK307 pKa = 10.55AMGDD311 pKa = 3.26DD312 pKa = 4.81SFEE315 pKa = 3.69IWFKK319 pKa = 11.17GLEE322 pKa = 4.08EE323 pKa = 4.1YY324 pKa = 10.04LGKK327 pKa = 9.87MGHH330 pKa = 4.88TVKK333 pKa = 9.93MCVQRR338 pKa = 11.84PGLVGFEE345 pKa = 4.36FCSQVFLGLGIAYY358 pKa = 8.89PVDD361 pKa = 4.28FSKK364 pKa = 9.95TLYY367 pKa = 10.33RR368 pKa = 11.84FLSHH372 pKa = 6.97HH373 pKa = 6.53PADD376 pKa = 4.18PKK378 pKa = 9.52YY379 pKa = 10.63SEE381 pKa = 4.15YY382 pKa = 10.34RR383 pKa = 11.84AQLMYY388 pKa = 10.46YY389 pKa = 10.03FRR391 pKa = 11.84HH392 pKa = 6.12LPSSTLQKK400 pKa = 10.56VIRR403 pKa = 11.84LAGARR408 pKa = 11.84VEE410 pKa = 4.29RR411 pKa = 11.84AQKK414 pKa = 10.61LATSSNN420 pKa = 3.6
MM1 pKa = 8.48DD2 pKa = 4.88SGLYY6 pKa = 9.61PKK8 pKa = 9.96TGAPTWFYY16 pKa = 10.36GWSKK20 pKa = 9.92QRR22 pKa = 11.84EE23 pKa = 4.4GGFSDD28 pKa = 4.43FGSEE32 pKa = 3.88IFRR35 pKa = 11.84LRR37 pKa = 11.84ALPPLKK43 pKa = 10.79AEE45 pKa = 3.94IFRR48 pKa = 11.84GIRR51 pKa = 11.84EE52 pKa = 4.03VVEE55 pKa = 3.84YY56 pKa = 10.82NITGDD61 pKa = 3.62SHH63 pKa = 7.4PGYY66 pKa = 8.99PWCKK70 pKa = 9.89LGSDD74 pKa = 3.67NKK76 pKa = 10.93AVLTGFGDD84 pKa = 4.55LIWDD88 pKa = 4.0EE89 pKa = 4.01VAKK92 pKa = 10.62RR93 pKa = 11.84FNNMLGYY100 pKa = 10.8GDD102 pKa = 5.99AIFSMTPSEE111 pKa = 4.05LVQNGICDD119 pKa = 3.58AVKK122 pKa = 10.72VFIKK126 pKa = 10.35QEE128 pKa = 3.77PHH130 pKa = 6.4SLEE133 pKa = 3.93KK134 pKa = 11.1VNAGRR139 pKa = 11.84LRR141 pKa = 11.84IIAAVGLVDD150 pKa = 4.84QIVTRR155 pKa = 11.84LLCMKK160 pKa = 10.14QNNAEE165 pKa = 3.86IDD167 pKa = 3.68CWEE170 pKa = 4.5SCPSAPGMGLNDD182 pKa = 3.31EE183 pKa = 4.73GLRR186 pKa = 11.84TLYY189 pKa = 9.67STAQVMAEE197 pKa = 4.36HH198 pKa = 6.23GTICEE203 pKa = 4.16TDD205 pKa = 2.74ISGWDD210 pKa = 3.21WSVQQWEE217 pKa = 4.41LDD219 pKa = 3.01SDD221 pKa = 3.66ARR223 pKa = 11.84LRR225 pKa = 11.84TQLAGEE231 pKa = 4.39EE232 pKa = 3.81IGGYY236 pKa = 10.65LNFFLRR242 pKa = 11.84VHH244 pKa = 6.88AYY246 pKa = 9.62VVGHH250 pKa = 5.83SVFVMPDD257 pKa = 3.08GEE259 pKa = 4.37MLEE262 pKa = 4.14QTVPGGQLSGDD273 pKa = 4.01YY274 pKa = 8.36NTSSSNSRR282 pKa = 11.84MRR284 pKa = 11.84VIATMFARR292 pKa = 11.84YY293 pKa = 8.52LAGQVSGFPLLGIKK307 pKa = 10.55AMGDD311 pKa = 3.26DD312 pKa = 4.81SFEE315 pKa = 3.69IWFKK319 pKa = 11.17GLEE322 pKa = 4.08EE323 pKa = 4.1YY324 pKa = 10.04LGKK327 pKa = 9.87MGHH330 pKa = 4.88TVKK333 pKa = 9.93MCVQRR338 pKa = 11.84PGLVGFEE345 pKa = 4.36FCSQVFLGLGIAYY358 pKa = 8.89PVDD361 pKa = 4.28FSKK364 pKa = 9.95TLYY367 pKa = 10.33RR368 pKa = 11.84FLSHH372 pKa = 6.97HH373 pKa = 6.53PADD376 pKa = 4.18PKK378 pKa = 9.52YY379 pKa = 10.63SEE381 pKa = 4.15YY382 pKa = 10.34RR383 pKa = 11.84AQLMYY388 pKa = 10.46YY389 pKa = 10.03FRR391 pKa = 11.84HH392 pKa = 6.12LPSSTLQKK400 pKa = 10.56VIRR403 pKa = 11.84LAGARR408 pKa = 11.84VEE410 pKa = 4.29RR411 pKa = 11.84AQKK414 pKa = 10.61LATSSNN420 pKa = 3.6
Molecular weight: 46.91 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q9YPD5|RDRP_MBVLF RNA-directed RNA polymerase OS=Mushroom bacilliform virus (isolate Australia/AUS LF-1) OX=650482 GN=ORF3 PE=3 SV=1
MM1 pKa = 7.69ANRR4 pKa = 11.84RR5 pKa = 11.84QSRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84GNKK13 pKa = 8.3NRR15 pKa = 11.84NSATVRR21 pKa = 11.84RR22 pKa = 11.84APPNRR27 pKa = 11.84ATQSSSGKK35 pKa = 9.61VKK37 pKa = 9.93FVKK40 pKa = 10.07WIAASPTKK48 pKa = 10.4LIPHH52 pKa = 6.7IGEE55 pKa = 4.31NEE57 pKa = 3.74TSYY60 pKa = 11.67GVLFDD65 pKa = 3.37ITGTTFPEE73 pKa = 4.63LSSLMSRR80 pKa = 11.84HH81 pKa = 4.32SRR83 pKa = 11.84YY84 pKa = 9.53RR85 pKa = 11.84VLSLGARR92 pKa = 11.84IVPYY96 pKa = 10.58DD97 pKa = 3.97PNCLGAHH104 pKa = 4.84SVKK107 pKa = 10.27VFAEE111 pKa = 4.09SVYY114 pKa = 10.99DD115 pKa = 3.82SSATPTVPSVHH126 pKa = 5.91YY127 pKa = 9.63LQSNGCRR134 pKa = 11.84VVPANKK140 pKa = 9.51QLSSPPSSDD149 pKa = 3.0VNKK152 pKa = 10.51YY153 pKa = 10.35YY154 pKa = 10.26EE155 pKa = 4.24ICSDD159 pKa = 3.65DD160 pKa = 3.82AVIGRR165 pKa = 11.84IMYY168 pKa = 9.46AWNGPGLSTARR179 pKa = 11.84VGFCSFEE186 pKa = 4.14VYY188 pKa = 11.01ADD190 pKa = 4.0LEE192 pKa = 4.15FDD194 pKa = 5.15GIRR197 pKa = 11.84EE198 pKa = 4.09
MM1 pKa = 7.69ANRR4 pKa = 11.84RR5 pKa = 11.84QSRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84GNKK13 pKa = 8.3NRR15 pKa = 11.84NSATVRR21 pKa = 11.84RR22 pKa = 11.84APPNRR27 pKa = 11.84ATQSSSGKK35 pKa = 9.61VKK37 pKa = 9.93FVKK40 pKa = 10.07WIAASPTKK48 pKa = 10.4LIPHH52 pKa = 6.7IGEE55 pKa = 4.31NEE57 pKa = 3.74TSYY60 pKa = 11.67GVLFDD65 pKa = 3.37ITGTTFPEE73 pKa = 4.63LSSLMSRR80 pKa = 11.84HH81 pKa = 4.32SRR83 pKa = 11.84YY84 pKa = 9.53RR85 pKa = 11.84VLSLGARR92 pKa = 11.84IVPYY96 pKa = 10.58DD97 pKa = 3.97PNCLGAHH104 pKa = 4.84SVKK107 pKa = 10.27VFAEE111 pKa = 4.09SVYY114 pKa = 10.99DD115 pKa = 3.82SSATPTVPSVHH126 pKa = 5.91YY127 pKa = 9.63LQSNGCRR134 pKa = 11.84VVPANKK140 pKa = 9.51QLSSPPSSDD149 pKa = 3.0VNKK152 pKa = 10.51YY153 pKa = 10.35YY154 pKa = 10.26EE155 pKa = 4.24ICSDD159 pKa = 3.65DD160 pKa = 3.82AVIGRR165 pKa = 11.84IMYY168 pKa = 9.46AWNGPGLSTARR179 pKa = 11.84VGFCSFEE186 pKa = 4.14VYY188 pKa = 11.01ADD190 pKa = 4.0LEE192 pKa = 4.15FDD194 pKa = 5.15GIRR197 pKa = 11.84EE198 pKa = 4.09
Molecular weight: 21.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1454 |
179 |
657 |
363.5 |
40.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.121 ± 0.586 | 2.338 ± 0.437 |
4.47 ± 0.217 | 5.983 ± 0.833 |
4.402 ± 0.334 | 7.497 ± 1.024 |
1.788 ± 0.464 | 4.195 ± 0.257 |
5.089 ± 0.579 | 9.216 ± 0.925 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.82 ± 0.381 | 3.508 ± 0.697 |
4.952 ± 0.519 | 3.164 ± 0.587 |
6.19 ± 0.699 | 9.422 ± 1.295 |
5.433 ± 0.728 | 8.391 ± 0.679 |
2.201 ± 0.393 | 2.82 ± 0.739 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
