
Celeribacter phage P12053L
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Zobellviridae; Cobavirinae; Siovirus; Celeribacter virus P12053L
Average proteome isoelectric point is 5.8
Get precalculated fractions of proteins
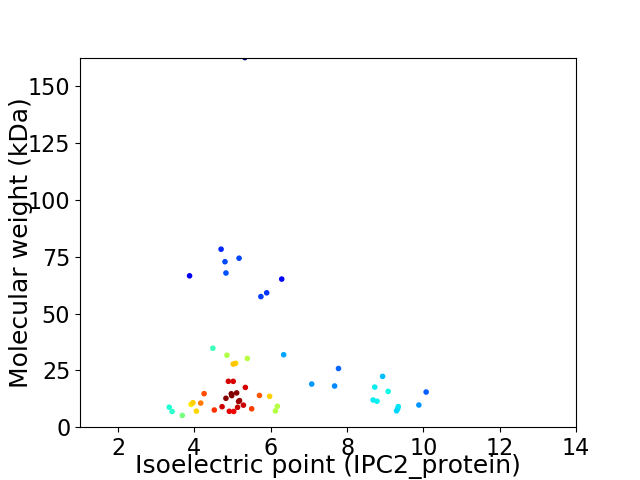
Virtual 2D-PAGE plot for 56 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I6S6K3|I6S6K3_9CAUD Putative phage tail fiber protein OS=Celeribacter phage P12053L OX=1197951 GN=P12053L_55 PE=4 SV=1
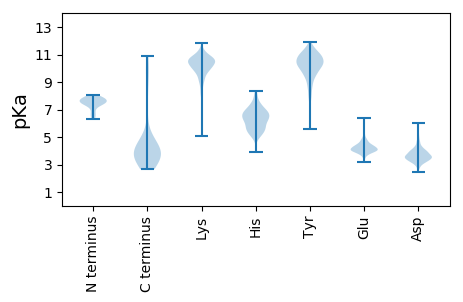
MM1 pKa = 7.59AKK3 pKa = 10.07KK4 pKa = 8.81PTVTTLQSGFNSTEE18 pKa = 3.77VLNSNFEE25 pKa = 4.19KK26 pKa = 10.87LRR28 pKa = 11.84DD29 pKa = 3.79AFDD32 pKa = 3.51NTLSLDD38 pKa = 3.47GSTPNAMEE46 pKa = 5.54ADD48 pKa = 3.65LDD50 pKa = 4.45LNGNNLIGAVGLLVNGTDD68 pKa = 3.62YY69 pKa = 11.12LAEE72 pKa = 4.04VEE74 pKa = 4.24AAKK77 pKa = 10.41AAALVAQAAAEE88 pKa = 4.08TAEE91 pKa = 4.5TNAEE95 pKa = 4.1TAEE98 pKa = 4.44TNAEE102 pKa = 4.16TAEE105 pKa = 4.04TAAASSASAASTSATNAATSATAASLSEE133 pKa = 3.83TAAASSEE140 pKa = 4.29ANTSASEE147 pKa = 4.06TAAGLSEE154 pKa = 4.03TAAATSEE161 pKa = 4.02TAAALSEE168 pKa = 4.36ANASASATAAATSATNASTSEE189 pKa = 4.07TNAAASATEE198 pKa = 3.99AATSEE203 pKa = 4.32ANAATSATNASTSEE217 pKa = 4.12TNAATSATAALASQTAAATSEE238 pKa = 4.72SNASTSAANAATSEE252 pKa = 4.61TNASTSASTASTKK265 pKa = 9.68ATEE268 pKa = 4.37AATSATNASTSEE280 pKa = 4.12TNAATSATSAATSATSAQASKK301 pKa = 10.94DD302 pKa = 3.5AALAALDD309 pKa = 4.23SFDD312 pKa = 5.84DD313 pKa = 4.23RR314 pKa = 11.84YY315 pKa = 10.94LGQKK319 pKa = 8.42ATDD322 pKa = 3.64PTLDD326 pKa = 3.42NDD328 pKa = 3.56GDD330 pKa = 4.24ALVTGALYY338 pKa = 10.63FNTADD343 pKa = 3.92DD344 pKa = 3.68VMKK347 pKa = 10.64VYY349 pKa = 10.56EE350 pKa = 4.21GSVWVAAYY358 pKa = 10.26ASLSGALLSANSLSDD373 pKa = 3.66VLNAAAARR381 pKa = 11.84TNLGLGTAATTASTDD396 pKa = 3.38YY397 pKa = 9.88ATAAQGSLADD407 pKa = 4.0SALQSGDD414 pKa = 3.82NISALTNDD422 pKa = 3.7AGYY425 pKa = 6.08TTNVGDD431 pKa = 3.89ITNVSAGTGLSGGGASGTVTISHH454 pKa = 7.61ADD456 pKa = 3.35TSSQGSVNNSGATVIQDD473 pKa = 3.54VTLDD477 pKa = 3.63TYY479 pKa = 11.63GHH481 pKa = 5.06VTGLASKK488 pKa = 8.75TLTAADD494 pKa = 3.44VGAITGNQTITLSGDD509 pKa = 3.19VSGSGTTSIVVTVADD524 pKa = 4.61DD525 pKa = 3.6SHH527 pKa = 7.82NHH529 pKa = 5.91VISNVDD535 pKa = 3.46GLQTALDD542 pKa = 3.83AKK544 pKa = 10.07LASSSYY550 pKa = 8.92TASDD554 pKa = 3.35VLTKK558 pKa = 10.85VKK560 pKa = 9.75TVDD563 pKa = 3.49GSGSGLDD570 pKa = 3.96ADD572 pKa = 5.38LLDD575 pKa = 4.65GQHH578 pKa = 5.85GSYY581 pKa = 10.51YY582 pKa = 10.04YY583 pKa = 10.89SPSNPPAASAPSSAQVGSATAGLSVGAVGTYY614 pKa = 10.32AYY616 pKa = 10.46LYY618 pKa = 8.5YY619 pKa = 10.53YY620 pKa = 9.91PRR622 pKa = 11.84TTTSPGTTRR631 pKa = 11.84AGSDD635 pKa = 2.95LRR637 pKa = 11.84YY638 pKa = 9.69ANGYY642 pKa = 8.54AAHH645 pKa = 6.57TGYY648 pKa = 9.18TGGGTPSGTWRR659 pKa = 11.84CMGYY663 pKa = 9.72IFDD666 pKa = 4.01QAGGTGTDD674 pKa = 3.37TTTVWLRR681 pKa = 11.84ISS683 pKa = 3.26
MM1 pKa = 7.59AKK3 pKa = 10.07KK4 pKa = 8.81PTVTTLQSGFNSTEE18 pKa = 3.77VLNSNFEE25 pKa = 4.19KK26 pKa = 10.87LRR28 pKa = 11.84DD29 pKa = 3.79AFDD32 pKa = 3.51NTLSLDD38 pKa = 3.47GSTPNAMEE46 pKa = 5.54ADD48 pKa = 3.65LDD50 pKa = 4.45LNGNNLIGAVGLLVNGTDD68 pKa = 3.62YY69 pKa = 11.12LAEE72 pKa = 4.04VEE74 pKa = 4.24AAKK77 pKa = 10.41AAALVAQAAAEE88 pKa = 4.08TAEE91 pKa = 4.5TNAEE95 pKa = 4.1TAEE98 pKa = 4.44TNAEE102 pKa = 4.16TAEE105 pKa = 4.04TAAASSASAASTSATNAATSATAASLSEE133 pKa = 3.83TAAASSEE140 pKa = 4.29ANTSASEE147 pKa = 4.06TAAGLSEE154 pKa = 4.03TAAATSEE161 pKa = 4.02TAAALSEE168 pKa = 4.36ANASASATAAATSATNASTSEE189 pKa = 4.07TNAAASATEE198 pKa = 3.99AATSEE203 pKa = 4.32ANAATSATNASTSEE217 pKa = 4.12TNAATSATAALASQTAAATSEE238 pKa = 4.72SNASTSAANAATSEE252 pKa = 4.61TNASTSASTASTKK265 pKa = 9.68ATEE268 pKa = 4.37AATSATNASTSEE280 pKa = 4.12TNAATSATSAATSATSAQASKK301 pKa = 10.94DD302 pKa = 3.5AALAALDD309 pKa = 4.23SFDD312 pKa = 5.84DD313 pKa = 4.23RR314 pKa = 11.84YY315 pKa = 10.94LGQKK319 pKa = 8.42ATDD322 pKa = 3.64PTLDD326 pKa = 3.42NDD328 pKa = 3.56GDD330 pKa = 4.24ALVTGALYY338 pKa = 10.63FNTADD343 pKa = 3.92DD344 pKa = 3.68VMKK347 pKa = 10.64VYY349 pKa = 10.56EE350 pKa = 4.21GSVWVAAYY358 pKa = 10.26ASLSGALLSANSLSDD373 pKa = 3.66VLNAAAARR381 pKa = 11.84TNLGLGTAATTASTDD396 pKa = 3.38YY397 pKa = 9.88ATAAQGSLADD407 pKa = 4.0SALQSGDD414 pKa = 3.82NISALTNDD422 pKa = 3.7AGYY425 pKa = 6.08TTNVGDD431 pKa = 3.89ITNVSAGTGLSGGGASGTVTISHH454 pKa = 7.61ADD456 pKa = 3.35TSSQGSVNNSGATVIQDD473 pKa = 3.54VTLDD477 pKa = 3.63TYY479 pKa = 11.63GHH481 pKa = 5.06VTGLASKK488 pKa = 8.75TLTAADD494 pKa = 3.44VGAITGNQTITLSGDD509 pKa = 3.19VSGSGTTSIVVTVADD524 pKa = 4.61DD525 pKa = 3.6SHH527 pKa = 7.82NHH529 pKa = 5.91VISNVDD535 pKa = 3.46GLQTALDD542 pKa = 3.83AKK544 pKa = 10.07LASSSYY550 pKa = 8.92TASDD554 pKa = 3.35VLTKK558 pKa = 10.85VKK560 pKa = 9.75TVDD563 pKa = 3.49GSGSGLDD570 pKa = 3.96ADD572 pKa = 5.38LLDD575 pKa = 4.65GQHH578 pKa = 5.85GSYY581 pKa = 10.51YY582 pKa = 10.04YY583 pKa = 10.89SPSNPPAASAPSSAQVGSATAGLSVGAVGTYY614 pKa = 10.32AYY616 pKa = 10.46LYY618 pKa = 8.5YY619 pKa = 10.53YY620 pKa = 9.91PRR622 pKa = 11.84TTTSPGTTRR631 pKa = 11.84AGSDD635 pKa = 2.95LRR637 pKa = 11.84YY638 pKa = 9.69ANGYY642 pKa = 8.54AAHH645 pKa = 6.57TGYY648 pKa = 9.18TGGGTPSGTWRR659 pKa = 11.84CMGYY663 pKa = 9.72IFDD666 pKa = 4.01QAGGTGTDD674 pKa = 3.37TTTVWLRR681 pKa = 11.84ISS683 pKa = 3.26
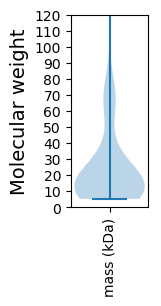
Molecular weight: 66.57 kDa
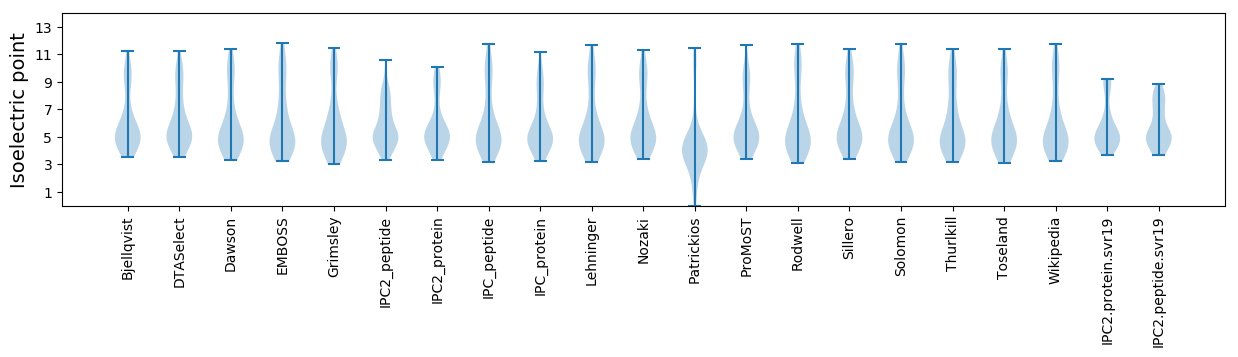
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I6RT32|I6RT32_9CAUD Uncharacterized protein OS=Celeribacter phage P12053L OX=1197951 GN=P12053L_48 PE=4 SV=1
MM1 pKa = 7.87AIFTAIGAALTAATGAAAFTAGVATVGAVVGVGGTIASGKK41 pKa = 8.38AARR44 pKa = 11.84RR45 pKa = 11.84AASAQQKK52 pKa = 9.68SQDD55 pKa = 3.76LQAKK59 pKa = 6.99RR60 pKa = 11.84QRR62 pKa = 11.84RR63 pKa = 11.84AAIRR67 pKa = 11.84SNILASARR75 pKa = 11.84ARR77 pKa = 11.84ASAQSAGTAQSSGLSGAIGAGRR99 pKa = 11.84SQLGAEE105 pKa = 4.56LGFGTQMSGLSANISKK121 pKa = 10.64FNMQAQTYY129 pKa = 10.35GDD131 pKa = 3.44IAKK134 pKa = 10.27LGFSAAGNAPALGGYY149 pKa = 10.03AEE151 pKa = 4.58EE152 pKa = 4.65FGLFNKK158 pKa = 10.19
MM1 pKa = 7.87AIFTAIGAALTAATGAAAFTAGVATVGAVVGVGGTIASGKK41 pKa = 8.38AARR44 pKa = 11.84RR45 pKa = 11.84AASAQQKK52 pKa = 9.68SQDD55 pKa = 3.76LQAKK59 pKa = 6.99RR60 pKa = 11.84QRR62 pKa = 11.84RR63 pKa = 11.84AAIRR67 pKa = 11.84SNILASARR75 pKa = 11.84ARR77 pKa = 11.84ASAQSAGTAQSSGLSGAIGAGRR99 pKa = 11.84SQLGAEE105 pKa = 4.56LGFGTQMSGLSANISKK121 pKa = 10.64FNMQAQTYY129 pKa = 10.35GDD131 pKa = 3.44IAKK134 pKa = 10.27LGFSAAGNAPALGGYY149 pKa = 10.03AEE151 pKa = 4.58EE152 pKa = 4.65FGLFNKK158 pKa = 10.19
Molecular weight: 15.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12540 |
46 |
1487 |
223.9 |
24.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.123 ± 0.833 | 0.949 ± 0.192 |
6.818 ± 0.212 | 7.073 ± 0.28 |
3.596 ± 0.224 | 7.121 ± 0.301 |
1.595 ± 0.167 | 4.777 ± 0.308 |
6.643 ± 0.442 | 8.094 ± 0.182 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.576 ± 0.167 | 4.609 ± 0.195 |
3.246 ± 0.179 | 3.517 ± 0.204 |
4.617 ± 0.321 | 6.89 ± 0.499 |
6.834 ± 0.516 | 6.722 ± 0.278 |
1.38 ± 0.18 | 3.82 ± 0.243 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
