
Beihai sobemo-like virus 24
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.59
Get precalculated fractions of proteins
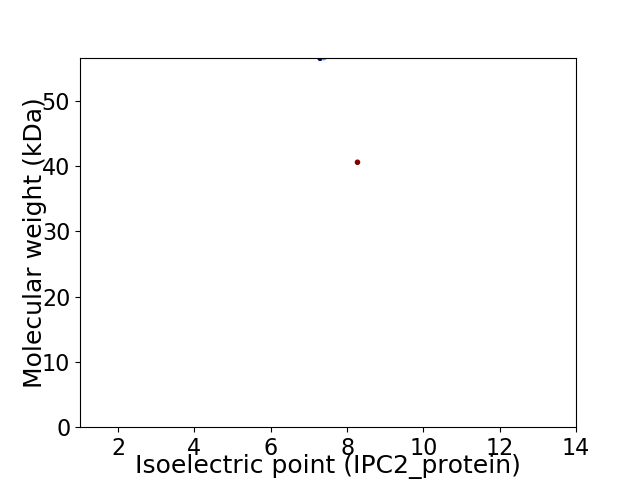
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KE68|A0A1L3KE68_9VIRU RNA-directed RNA polymerase OS=Beihai sobemo-like virus 24 OX=1922696 PE=4 SV=1
MM1 pKa = 7.54EE2 pKa = 6.19PIEE5 pKa = 4.77MITTVLGTLVLWEE18 pKa = 4.04IVRR21 pKa = 11.84SLFRR25 pKa = 11.84LLAQRR30 pKa = 11.84FRR32 pKa = 11.84RR33 pKa = 11.84VNVTGKK39 pKa = 8.07TLEE42 pKa = 4.38DD43 pKa = 3.51YY44 pKa = 11.04QLIGEE49 pKa = 4.53DD50 pKa = 3.75VACRR54 pKa = 11.84RR55 pKa = 11.84VYY57 pKa = 11.3KK58 pKa = 8.36MTKK61 pKa = 9.83SDD63 pKa = 3.74FQFEE67 pKa = 4.71SIRR70 pKa = 11.84KK71 pKa = 8.21NSKK74 pKa = 10.42LEE76 pKa = 3.94EE77 pKa = 4.25TKK79 pKa = 10.62SIHH82 pKa = 5.8QDD84 pKa = 3.05QVALLDD90 pKa = 3.95KK91 pKa = 10.22EE92 pKa = 4.72GTFIGYY98 pKa = 9.01GIRR101 pKa = 11.84FEE103 pKa = 5.17AGLVLPHH110 pKa = 6.67HH111 pKa = 6.99VWQCARR117 pKa = 11.84NGFLIGPTGKK127 pKa = 9.68QVAMEE132 pKa = 4.42GKK134 pKa = 8.51LTIISTDD141 pKa = 3.53LAIVDD146 pKa = 4.51IPNKK150 pKa = 9.68SWSTLGVKK158 pKa = 7.56VTKK161 pKa = 9.78PIPVAGNTFAEE172 pKa = 4.49ASGNPLLDD180 pKa = 4.89DD181 pKa = 4.13GLVRR185 pKa = 11.84LQGHH189 pKa = 6.44RR190 pKa = 11.84SFGYY194 pKa = 10.49LEE196 pKa = 4.22PASFGMLHH204 pKa = 6.14YY205 pKa = 10.69GGSTRR210 pKa = 11.84RR211 pKa = 11.84GMSGCGYY218 pKa = 10.59YY219 pKa = 10.64VGDD222 pKa = 4.32KK223 pKa = 10.79LAGLHH228 pKa = 6.32IGGGLVNHH236 pKa = 7.49GYY238 pKa = 9.8SAAFIQAITSHH249 pKa = 6.39GEE251 pKa = 3.79STEE254 pKa = 5.01DD255 pKa = 3.59YY256 pKa = 11.23LLDD259 pKa = 3.45LVKK262 pKa = 10.38RR263 pKa = 11.84RR264 pKa = 11.84KK265 pKa = 9.77RR266 pKa = 11.84GLDD269 pKa = 3.23YY270 pKa = 11.04RR271 pKa = 11.84RR272 pKa = 11.84VPGLLGEE279 pKa = 4.46VAVCLPGGRR288 pKa = 11.84FTIMEE293 pKa = 4.33EE294 pKa = 3.83EE295 pKa = 4.37DD296 pKa = 3.65ALEE299 pKa = 4.35LGILGDD305 pKa = 4.34EE306 pKa = 4.48LFKK309 pKa = 11.31SPEE312 pKa = 4.01GAVSYY317 pKa = 10.5EE318 pKa = 3.95DD319 pKa = 3.98QVLKK323 pKa = 10.14MKK325 pKa = 10.6NVEE328 pKa = 4.15IPSDD332 pKa = 3.35PTEE335 pKa = 3.57AHH337 pKa = 5.94FVKK340 pKa = 10.47EE341 pKa = 4.0VLDD344 pKa = 4.19AGGEE348 pKa = 4.1AEE350 pKa = 4.78TIGDD354 pKa = 4.37LDD356 pKa = 3.97SVFQKK361 pKa = 10.96RR362 pKa = 11.84LGDD365 pKa = 3.56QAQKK369 pKa = 10.94LRR371 pKa = 11.84DD372 pKa = 3.69EE373 pKa = 4.7LKK375 pKa = 10.63AIQEE379 pKa = 4.02QRR381 pKa = 11.84AKK383 pKa = 10.63EE384 pKa = 4.15SVRR387 pKa = 11.84PKK389 pKa = 10.07VQPSDD394 pKa = 3.4TTLLEE399 pKa = 4.19SQISSLKK406 pKa = 9.62EE407 pKa = 3.3QLKK410 pKa = 10.62AIRR413 pKa = 11.84LEE415 pKa = 4.22DD416 pKa = 3.27QQQRR420 pKa = 11.84EE421 pKa = 4.19KK422 pKa = 10.9QKK424 pKa = 10.95QEE426 pKa = 3.64VQADD430 pKa = 3.91RR431 pKa = 11.84EE432 pKa = 4.44YY433 pKa = 10.92QQSLLEE439 pKa = 4.23KK440 pKa = 10.1KK441 pKa = 10.13KK442 pKa = 10.4QLEE445 pKa = 4.21KK446 pKa = 10.63EE447 pKa = 4.11LASVSVLASSRR458 pKa = 11.84NYY460 pKa = 8.75LTKK463 pKa = 10.36QSKK466 pKa = 10.59SSLKK470 pKa = 10.16RR471 pKa = 11.84EE472 pKa = 3.68KK473 pKa = 10.83ARR475 pKa = 11.84AKK477 pKa = 10.5KK478 pKa = 8.36MQEE481 pKa = 3.96EE482 pKa = 4.06AQHH485 pKa = 6.26AKK487 pKa = 9.22VAKK490 pKa = 9.77EE491 pKa = 3.76LEE493 pKa = 4.35EE494 pKa = 3.96LRR496 pKa = 11.84KK497 pKa = 9.69LVPTLGTKK505 pKa = 8.9SS506 pKa = 3.19
MM1 pKa = 7.54EE2 pKa = 6.19PIEE5 pKa = 4.77MITTVLGTLVLWEE18 pKa = 4.04IVRR21 pKa = 11.84SLFRR25 pKa = 11.84LLAQRR30 pKa = 11.84FRR32 pKa = 11.84RR33 pKa = 11.84VNVTGKK39 pKa = 8.07TLEE42 pKa = 4.38DD43 pKa = 3.51YY44 pKa = 11.04QLIGEE49 pKa = 4.53DD50 pKa = 3.75VACRR54 pKa = 11.84RR55 pKa = 11.84VYY57 pKa = 11.3KK58 pKa = 8.36MTKK61 pKa = 9.83SDD63 pKa = 3.74FQFEE67 pKa = 4.71SIRR70 pKa = 11.84KK71 pKa = 8.21NSKK74 pKa = 10.42LEE76 pKa = 3.94EE77 pKa = 4.25TKK79 pKa = 10.62SIHH82 pKa = 5.8QDD84 pKa = 3.05QVALLDD90 pKa = 3.95KK91 pKa = 10.22EE92 pKa = 4.72GTFIGYY98 pKa = 9.01GIRR101 pKa = 11.84FEE103 pKa = 5.17AGLVLPHH110 pKa = 6.67HH111 pKa = 6.99VWQCARR117 pKa = 11.84NGFLIGPTGKK127 pKa = 9.68QVAMEE132 pKa = 4.42GKK134 pKa = 8.51LTIISTDD141 pKa = 3.53LAIVDD146 pKa = 4.51IPNKK150 pKa = 9.68SWSTLGVKK158 pKa = 7.56VTKK161 pKa = 9.78PIPVAGNTFAEE172 pKa = 4.49ASGNPLLDD180 pKa = 4.89DD181 pKa = 4.13GLVRR185 pKa = 11.84LQGHH189 pKa = 6.44RR190 pKa = 11.84SFGYY194 pKa = 10.49LEE196 pKa = 4.22PASFGMLHH204 pKa = 6.14YY205 pKa = 10.69GGSTRR210 pKa = 11.84RR211 pKa = 11.84GMSGCGYY218 pKa = 10.59YY219 pKa = 10.64VGDD222 pKa = 4.32KK223 pKa = 10.79LAGLHH228 pKa = 6.32IGGGLVNHH236 pKa = 7.49GYY238 pKa = 9.8SAAFIQAITSHH249 pKa = 6.39GEE251 pKa = 3.79STEE254 pKa = 5.01DD255 pKa = 3.59YY256 pKa = 11.23LLDD259 pKa = 3.45LVKK262 pKa = 10.38RR263 pKa = 11.84RR264 pKa = 11.84KK265 pKa = 9.77RR266 pKa = 11.84GLDD269 pKa = 3.23YY270 pKa = 11.04RR271 pKa = 11.84RR272 pKa = 11.84VPGLLGEE279 pKa = 4.46VAVCLPGGRR288 pKa = 11.84FTIMEE293 pKa = 4.33EE294 pKa = 3.83EE295 pKa = 4.37DD296 pKa = 3.65ALEE299 pKa = 4.35LGILGDD305 pKa = 4.34EE306 pKa = 4.48LFKK309 pKa = 11.31SPEE312 pKa = 4.01GAVSYY317 pKa = 10.5EE318 pKa = 3.95DD319 pKa = 3.98QVLKK323 pKa = 10.14MKK325 pKa = 10.6NVEE328 pKa = 4.15IPSDD332 pKa = 3.35PTEE335 pKa = 3.57AHH337 pKa = 5.94FVKK340 pKa = 10.47EE341 pKa = 4.0VLDD344 pKa = 4.19AGGEE348 pKa = 4.1AEE350 pKa = 4.78TIGDD354 pKa = 4.37LDD356 pKa = 3.97SVFQKK361 pKa = 10.96RR362 pKa = 11.84LGDD365 pKa = 3.56QAQKK369 pKa = 10.94LRR371 pKa = 11.84DD372 pKa = 3.69EE373 pKa = 4.7LKK375 pKa = 10.63AIQEE379 pKa = 4.02QRR381 pKa = 11.84AKK383 pKa = 10.63EE384 pKa = 4.15SVRR387 pKa = 11.84PKK389 pKa = 10.07VQPSDD394 pKa = 3.4TTLLEE399 pKa = 4.19SQISSLKK406 pKa = 9.62EE407 pKa = 3.3QLKK410 pKa = 10.62AIRR413 pKa = 11.84LEE415 pKa = 4.22DD416 pKa = 3.27QQQRR420 pKa = 11.84EE421 pKa = 4.19KK422 pKa = 10.9QKK424 pKa = 10.95QEE426 pKa = 3.64VQADD430 pKa = 3.91RR431 pKa = 11.84EE432 pKa = 4.44YY433 pKa = 10.92QQSLLEE439 pKa = 4.23KK440 pKa = 10.1KK441 pKa = 10.13KK442 pKa = 10.4QLEE445 pKa = 4.21KK446 pKa = 10.63EE447 pKa = 4.11LASVSVLASSRR458 pKa = 11.84NYY460 pKa = 8.75LTKK463 pKa = 10.36QSKK466 pKa = 10.59SSLKK470 pKa = 10.16RR471 pKa = 11.84EE472 pKa = 3.68KK473 pKa = 10.83ARR475 pKa = 11.84AKK477 pKa = 10.5KK478 pKa = 8.36MQEE481 pKa = 3.96EE482 pKa = 4.06AQHH485 pKa = 6.26AKK487 pKa = 9.22VAKK490 pKa = 9.77EE491 pKa = 3.76LEE493 pKa = 4.35EE494 pKa = 3.96LRR496 pKa = 11.84KK497 pKa = 9.69LVPTLGTKK505 pKa = 8.9SS506 pKa = 3.19
Molecular weight: 56.49 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KE68|A0A1L3KE68_9VIRU RNA-directed RNA polymerase OS=Beihai sobemo-like virus 24 OX=1922696 PE=4 SV=1
MM1 pKa = 7.84DD2 pKa = 3.69WDD4 pKa = 3.93IYY6 pKa = 11.13RR7 pKa = 11.84KK8 pKa = 9.3ILEE11 pKa = 4.77EE12 pKa = 4.18IDD14 pKa = 3.65PKK16 pKa = 10.89KK17 pKa = 11.0SPGYY21 pKa = 9.12PFKK24 pKa = 11.05AWGITTNKK32 pKa = 10.02QILDD36 pKa = 3.74SSKK39 pKa = 10.37MLVQLYY45 pKa = 10.58CMVRR49 pKa = 11.84QRR51 pKa = 11.84LDD53 pKa = 2.9QLARR57 pKa = 11.84GRR59 pKa = 11.84VGGDD63 pKa = 4.03DD64 pKa = 3.07INLFIKK70 pKa = 10.36QEE72 pKa = 3.82PHH74 pKa = 6.49KK75 pKa = 10.38KK76 pKa = 9.98SKK78 pKa = 10.55AAAKK82 pKa = 9.68RR83 pKa = 11.84WRR85 pKa = 11.84LISGVGLVDD94 pKa = 4.32NIVDD98 pKa = 3.67RR99 pKa = 11.84FLFGGLLEE107 pKa = 4.51KK108 pKa = 10.5VDD110 pKa = 4.22YY111 pKa = 10.09LAKK114 pKa = 10.39RR115 pKa = 11.84MKK117 pKa = 10.34IPILAGWVPWGGGYY131 pKa = 9.71RR132 pKa = 11.84ALANTIQEE140 pKa = 4.26PQSCDD145 pKa = 2.81KK146 pKa = 11.22SAWDD150 pKa = 3.26WTMQGWIVQGFQQILRR166 pKa = 11.84TIQCNPDD173 pKa = 2.56WSLAIDD179 pKa = 3.65ARR181 pKa = 11.84LATLFNASVFRR192 pKa = 11.84VGEE195 pKa = 3.88RR196 pKa = 11.84RR197 pKa = 11.84FKK199 pKa = 10.45QNVMGIMKK207 pKa = 10.21SGFLGTIVFNSIGQMLIHH225 pKa = 6.59HH226 pKa = 7.07LSLLRR231 pKa = 11.84GKK233 pKa = 10.15GKK235 pKa = 9.88RR236 pKa = 11.84LPYY239 pKa = 10.1FVVGDD244 pKa = 3.87DD245 pKa = 3.79MVTEE249 pKa = 4.35KK250 pKa = 9.49QTPAYY255 pKa = 8.02WSAVEE260 pKa = 4.18TSGCSLKK267 pKa = 10.49EE268 pKa = 3.95YY269 pKa = 9.64QEE271 pKa = 4.27KK272 pKa = 10.23PGYY275 pKa = 7.29PTEE278 pKa = 4.04FMGVKK283 pKa = 10.43FSEE286 pKa = 4.16NVILPAYY293 pKa = 8.33PDD295 pKa = 3.43KK296 pKa = 11.8NMFSLLIKK304 pKa = 10.17EE305 pKa = 4.14PEE307 pKa = 3.86ILKK310 pKa = 10.28EE311 pKa = 4.12ALRR314 pKa = 11.84CYY316 pKa = 9.96QLLYY320 pKa = 9.92SHH322 pKa = 7.06HH323 pKa = 7.2PLRR326 pKa = 11.84SVVADD331 pKa = 3.77LAKK334 pKa = 10.37AYY336 pKa = 8.38EE337 pKa = 4.23VQLPEE342 pKa = 4.31SEE344 pKa = 4.35VVSWVEE350 pKa = 3.75GWEE353 pKa = 4.34CAA355 pKa = 4.38
MM1 pKa = 7.84DD2 pKa = 3.69WDD4 pKa = 3.93IYY6 pKa = 11.13RR7 pKa = 11.84KK8 pKa = 9.3ILEE11 pKa = 4.77EE12 pKa = 4.18IDD14 pKa = 3.65PKK16 pKa = 10.89KK17 pKa = 11.0SPGYY21 pKa = 9.12PFKK24 pKa = 11.05AWGITTNKK32 pKa = 10.02QILDD36 pKa = 3.74SSKK39 pKa = 10.37MLVQLYY45 pKa = 10.58CMVRR49 pKa = 11.84QRR51 pKa = 11.84LDD53 pKa = 2.9QLARR57 pKa = 11.84GRR59 pKa = 11.84VGGDD63 pKa = 4.03DD64 pKa = 3.07INLFIKK70 pKa = 10.36QEE72 pKa = 3.82PHH74 pKa = 6.49KK75 pKa = 10.38KK76 pKa = 9.98SKK78 pKa = 10.55AAAKK82 pKa = 9.68RR83 pKa = 11.84WRR85 pKa = 11.84LISGVGLVDD94 pKa = 4.32NIVDD98 pKa = 3.67RR99 pKa = 11.84FLFGGLLEE107 pKa = 4.51KK108 pKa = 10.5VDD110 pKa = 4.22YY111 pKa = 10.09LAKK114 pKa = 10.39RR115 pKa = 11.84MKK117 pKa = 10.34IPILAGWVPWGGGYY131 pKa = 9.71RR132 pKa = 11.84ALANTIQEE140 pKa = 4.26PQSCDD145 pKa = 2.81KK146 pKa = 11.22SAWDD150 pKa = 3.26WTMQGWIVQGFQQILRR166 pKa = 11.84TIQCNPDD173 pKa = 2.56WSLAIDD179 pKa = 3.65ARR181 pKa = 11.84LATLFNASVFRR192 pKa = 11.84VGEE195 pKa = 3.88RR196 pKa = 11.84RR197 pKa = 11.84FKK199 pKa = 10.45QNVMGIMKK207 pKa = 10.21SGFLGTIVFNSIGQMLIHH225 pKa = 6.59HH226 pKa = 7.07LSLLRR231 pKa = 11.84GKK233 pKa = 10.15GKK235 pKa = 9.88RR236 pKa = 11.84LPYY239 pKa = 10.1FVVGDD244 pKa = 3.87DD245 pKa = 3.79MVTEE249 pKa = 4.35KK250 pKa = 9.49QTPAYY255 pKa = 8.02WSAVEE260 pKa = 4.18TSGCSLKK267 pKa = 10.49EE268 pKa = 3.95YY269 pKa = 9.64QEE271 pKa = 4.27KK272 pKa = 10.23PGYY275 pKa = 7.29PTEE278 pKa = 4.04FMGVKK283 pKa = 10.43FSEE286 pKa = 4.16NVILPAYY293 pKa = 8.33PDD295 pKa = 3.43KK296 pKa = 11.8NMFSLLIKK304 pKa = 10.17EE305 pKa = 4.14PEE307 pKa = 3.86ILKK310 pKa = 10.28EE311 pKa = 4.12ALRR314 pKa = 11.84CYY316 pKa = 9.96QLLYY320 pKa = 9.92SHH322 pKa = 7.06HH323 pKa = 7.2PLRR326 pKa = 11.84SVVADD331 pKa = 3.77LAKK334 pKa = 10.37AYY336 pKa = 8.38EE337 pKa = 4.23VQLPEE342 pKa = 4.31SEE344 pKa = 4.35VVSWVEE350 pKa = 3.75GWEE353 pKa = 4.34CAA355 pKa = 4.38
Molecular weight: 40.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
861 |
355 |
506 |
430.5 |
48.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.388 ± 0.115 | 1.161 ± 0.318 |
4.994 ± 0.046 | 7.666 ± 1.221 |
3.368 ± 0.346 | 8.13 ± 0.315 |
1.742 ± 0.201 | 5.459 ± 0.613 |
8.014 ± 0.245 | 11.034 ± 0.537 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.323 ± 0.466 | 2.207 ± 0.367 |
3.833 ± 0.575 | 5.575 ± 0.303 |
5.807 ± 0.274 | 6.388 ± 0.284 |
4.181 ± 0.651 | 6.969 ± 0.044 |
1.742 ± 0.985 | 3.02 ± 0.386 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
