
Norwalk virus (strain GI/Human/United States/Norwalk/1968) (Hu/NV/NV/1968/US)
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Picornavirales; Caliciviridae; Norovirus; Norwalk virus; Norovirus isolates
Average proteome isoelectric point is 6.91
Get precalculated fractions of proteins
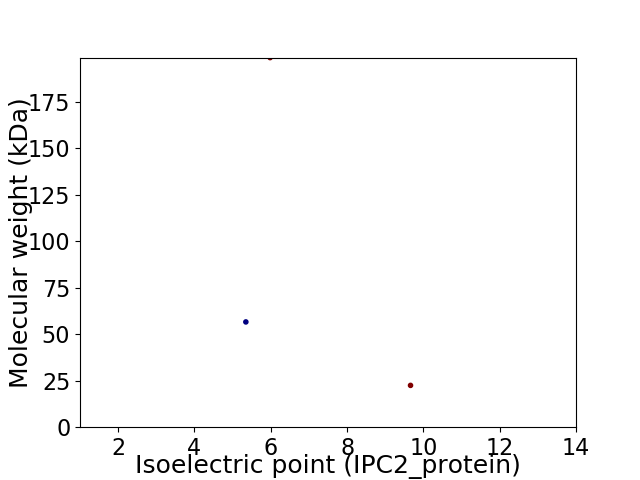
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q83885|VP2_NVN68 Protein VP2 OS=Norwalk virus (strain GI/Human/United States/Norwalk/1968) OX=524364 GN=ORF3 PE=3 SV=1
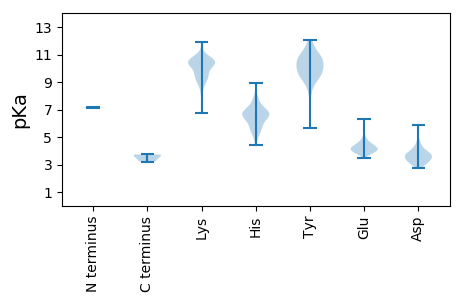
MM1 pKa = 7.14MMASKK6 pKa = 10.62DD7 pKa = 3.49ATSSVDD13 pKa = 3.33GASGAGQLVPEE24 pKa = 4.82VNASDD29 pKa = 4.55PLAMDD34 pKa = 4.29PVAGSSTAVATAGQVNPIDD53 pKa = 3.3PWIINNFVQAPQGEE67 pKa = 4.72FTISPNNTPGDD78 pKa = 3.77VLFDD82 pKa = 4.42LSLGPHH88 pKa = 6.92LNPFLLHH95 pKa = 6.7LSQMYY100 pKa = 10.11NGWVGNMRR108 pKa = 11.84VRR110 pKa = 11.84IMLAGNAFTAGKK122 pKa = 9.71IIVSCIPPGFGSHH135 pKa = 6.39NLTIAQATLFPHH147 pKa = 6.69VIADD151 pKa = 3.52VRR153 pKa = 11.84TLDD156 pKa = 4.47PIEE159 pKa = 4.53VPLEE163 pKa = 3.86DD164 pKa = 3.42VRR166 pKa = 11.84NVLFHH171 pKa = 6.97NNDD174 pKa = 3.29RR175 pKa = 11.84NQQTMRR181 pKa = 11.84LVCMLYY187 pKa = 9.52TPLRR191 pKa = 11.84TGGGTGDD198 pKa = 3.66SFVVAGRR205 pKa = 11.84VMTCPSPDD213 pKa = 3.49FNFLFLVPPTVEE225 pKa = 3.74QKK227 pKa = 8.96TRR229 pKa = 11.84PFTLPNLPLSSLSNSRR245 pKa = 11.84APLPISSMGISPDD258 pKa = 3.29NVQSVQFQNGRR269 pKa = 11.84CTLDD273 pKa = 3.03GRR275 pKa = 11.84LVGTTPVSLSHH286 pKa = 5.38VAKK289 pKa = 10.57IRR291 pKa = 11.84GTSNGTVINLTEE303 pKa = 4.18LDD305 pKa = 3.86GTPFHH310 pKa = 6.94PFEE313 pKa = 5.62GPAPIGFPDD322 pKa = 4.63LGGCDD327 pKa = 2.82WHH329 pKa = 8.91INMTQFGHH337 pKa = 6.56SSQTQYY343 pKa = 11.76DD344 pKa = 3.76VDD346 pKa = 3.97TTPDD350 pKa = 3.15TFVPHH355 pKa = 6.93LGSIQANGIGSGNYY369 pKa = 9.67VGVLSWISPPSHH381 pKa = 6.98PSGSQVDD388 pKa = 3.57LWKK391 pKa = 10.44IPNYY395 pKa = 10.07GSSITEE401 pKa = 4.01ATHH404 pKa = 6.83LAPSVYY410 pKa = 9.74PPGFGEE416 pKa = 4.08VLVFFMSKK424 pKa = 9.24MPGPGAYY431 pKa = 9.94NLPCLLPQEE440 pKa = 4.92YY441 pKa = 9.31ISHH444 pKa = 7.11LASEE448 pKa = 4.39QAPTVGEE455 pKa = 4.1AALLHH460 pKa = 6.16YY461 pKa = 10.43VDD463 pKa = 5.61PDD465 pKa = 3.24TGRR468 pKa = 11.84NLGEE472 pKa = 4.03FKK474 pKa = 10.59AYY476 pKa = 9.47PDD478 pKa = 3.78GFLTCVPNGASSGPQQLPINGVFVFVSWVSRR509 pKa = 11.84FYY511 pKa = 10.66QLKK514 pKa = 9.46PVGTASSARR523 pKa = 11.84GRR525 pKa = 11.84LGLRR529 pKa = 11.84RR530 pKa = 3.73
MM1 pKa = 7.14MMASKK6 pKa = 10.62DD7 pKa = 3.49ATSSVDD13 pKa = 3.33GASGAGQLVPEE24 pKa = 4.82VNASDD29 pKa = 4.55PLAMDD34 pKa = 4.29PVAGSSTAVATAGQVNPIDD53 pKa = 3.3PWIINNFVQAPQGEE67 pKa = 4.72FTISPNNTPGDD78 pKa = 3.77VLFDD82 pKa = 4.42LSLGPHH88 pKa = 6.92LNPFLLHH95 pKa = 6.7LSQMYY100 pKa = 10.11NGWVGNMRR108 pKa = 11.84VRR110 pKa = 11.84IMLAGNAFTAGKK122 pKa = 9.71IIVSCIPPGFGSHH135 pKa = 6.39NLTIAQATLFPHH147 pKa = 6.69VIADD151 pKa = 3.52VRR153 pKa = 11.84TLDD156 pKa = 4.47PIEE159 pKa = 4.53VPLEE163 pKa = 3.86DD164 pKa = 3.42VRR166 pKa = 11.84NVLFHH171 pKa = 6.97NNDD174 pKa = 3.29RR175 pKa = 11.84NQQTMRR181 pKa = 11.84LVCMLYY187 pKa = 9.52TPLRR191 pKa = 11.84TGGGTGDD198 pKa = 3.66SFVVAGRR205 pKa = 11.84VMTCPSPDD213 pKa = 3.49FNFLFLVPPTVEE225 pKa = 3.74QKK227 pKa = 8.96TRR229 pKa = 11.84PFTLPNLPLSSLSNSRR245 pKa = 11.84APLPISSMGISPDD258 pKa = 3.29NVQSVQFQNGRR269 pKa = 11.84CTLDD273 pKa = 3.03GRR275 pKa = 11.84LVGTTPVSLSHH286 pKa = 5.38VAKK289 pKa = 10.57IRR291 pKa = 11.84GTSNGTVINLTEE303 pKa = 4.18LDD305 pKa = 3.86GTPFHH310 pKa = 6.94PFEE313 pKa = 5.62GPAPIGFPDD322 pKa = 4.63LGGCDD327 pKa = 2.82WHH329 pKa = 8.91INMTQFGHH337 pKa = 6.56SSQTQYY343 pKa = 11.76DD344 pKa = 3.76VDD346 pKa = 3.97TTPDD350 pKa = 3.15TFVPHH355 pKa = 6.93LGSIQANGIGSGNYY369 pKa = 9.67VGVLSWISPPSHH381 pKa = 6.98PSGSQVDD388 pKa = 3.57LWKK391 pKa = 10.44IPNYY395 pKa = 10.07GSSITEE401 pKa = 4.01ATHH404 pKa = 6.83LAPSVYY410 pKa = 9.74PPGFGEE416 pKa = 4.08VLVFFMSKK424 pKa = 9.24MPGPGAYY431 pKa = 9.94NLPCLLPQEE440 pKa = 4.92YY441 pKa = 9.31ISHH444 pKa = 7.11LASEE448 pKa = 4.39QAPTVGEE455 pKa = 4.1AALLHH460 pKa = 6.16YY461 pKa = 10.43VDD463 pKa = 5.61PDD465 pKa = 3.24TGRR468 pKa = 11.84NLGEE472 pKa = 4.03FKK474 pKa = 10.59AYY476 pKa = 9.47PDD478 pKa = 3.78GFLTCVPNGASSGPQQLPINGVFVFVSWVSRR509 pKa = 11.84FYY511 pKa = 10.66QLKK514 pKa = 9.46PVGTASSARR523 pKa = 11.84GRR525 pKa = 11.84LGLRR529 pKa = 11.84RR530 pKa = 3.73
Molecular weight: 56.59 kDa
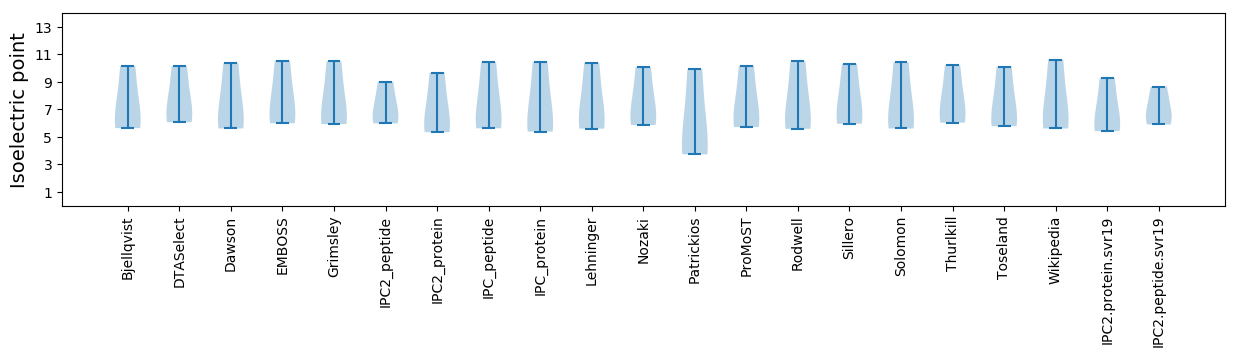
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q83885|VP2_NVN68 Protein VP2 OS=Norwalk virus (strain GI/Human/United States/Norwalk/1968) OX=524364 GN=ORF3 PE=3 SV=1
MM1 pKa = 7.14AQAIIGAIAASTAGSALGAGIQVGGEE27 pKa = 3.81AALQSQRR34 pKa = 11.84YY35 pKa = 5.62QQNLQLQEE43 pKa = 4.08NSFKK47 pKa = 10.49HH48 pKa = 5.32DD49 pKa = 3.36RR50 pKa = 11.84EE51 pKa = 4.22MIGYY55 pKa = 8.43QVEE58 pKa = 4.17ASNQLLAKK66 pKa = 10.25NLATRR71 pKa = 11.84YY72 pKa = 9.64SLLRR76 pKa = 11.84AGGLTSADD84 pKa = 3.32AARR87 pKa = 11.84SVAGAPVTRR96 pKa = 11.84IVDD99 pKa = 3.52WNGVRR104 pKa = 11.84VSAPEE109 pKa = 3.87SSATTLRR116 pKa = 11.84SGGFMSVPIPFASKK130 pKa = 9.85QKK132 pKa = 9.84QVQSSGISNPNYY144 pKa = 9.74SPSSISRR151 pKa = 11.84TTSWVEE157 pKa = 3.84SQNSSRR163 pKa = 11.84FGNLSPYY170 pKa = 9.62HH171 pKa = 7.12AEE173 pKa = 3.9ALNTVWLTPPGSTASSTLSSVPRR196 pKa = 11.84GYY198 pKa = 11.02FNTDD202 pKa = 3.16RR203 pKa = 11.84LPLFANNRR211 pKa = 11.84RR212 pKa = 3.65
MM1 pKa = 7.14AQAIIGAIAASTAGSALGAGIQVGGEE27 pKa = 3.81AALQSQRR34 pKa = 11.84YY35 pKa = 5.62QQNLQLQEE43 pKa = 4.08NSFKK47 pKa = 10.49HH48 pKa = 5.32DD49 pKa = 3.36RR50 pKa = 11.84EE51 pKa = 4.22MIGYY55 pKa = 8.43QVEE58 pKa = 4.17ASNQLLAKK66 pKa = 10.25NLATRR71 pKa = 11.84YY72 pKa = 9.64SLLRR76 pKa = 11.84AGGLTSADD84 pKa = 3.32AARR87 pKa = 11.84SVAGAPVTRR96 pKa = 11.84IVDD99 pKa = 3.52WNGVRR104 pKa = 11.84VSAPEE109 pKa = 3.87SSATTLRR116 pKa = 11.84SGGFMSVPIPFASKK130 pKa = 9.85QKK132 pKa = 9.84QVQSSGISNPNYY144 pKa = 9.74SPSSISRR151 pKa = 11.84TTSWVEE157 pKa = 3.84SQNSSRR163 pKa = 11.84FGNLSPYY170 pKa = 9.62HH171 pKa = 7.12AEE173 pKa = 3.9ALNTVWLTPPGSTASSTLSSVPRR196 pKa = 11.84GYY198 pKa = 11.02FNTDD202 pKa = 3.16RR203 pKa = 11.84LPLFANNRR211 pKa = 11.84RR212 pKa = 3.65
Molecular weight: 22.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2531 |
212 |
1789 |
843.7 |
92.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.744 ± 0.986 | 1.304 ± 0.277 |
5.334 ± 0.85 | 5.452 ± 1.459 |
3.635 ± 0.583 | 8.376 ± 0.582 |
2.252 ± 0.279 | 4.939 ± 0.258 |
4.82 ± 1.682 | 8.179 ± 0.359 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.726 ± 0.293 | 4.188 ± 0.839 |
6.519 ± 1.321 | 4.623 ± 0.411 |
5.255 ± 0.672 | 7.428 ± 1.766 |
6.282 ± 0.129 | 6.875 ± 0.576 |
1.58 ± 0.201 | 2.489 ± 0.167 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
