
Shahe picorna-like virus 11
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins
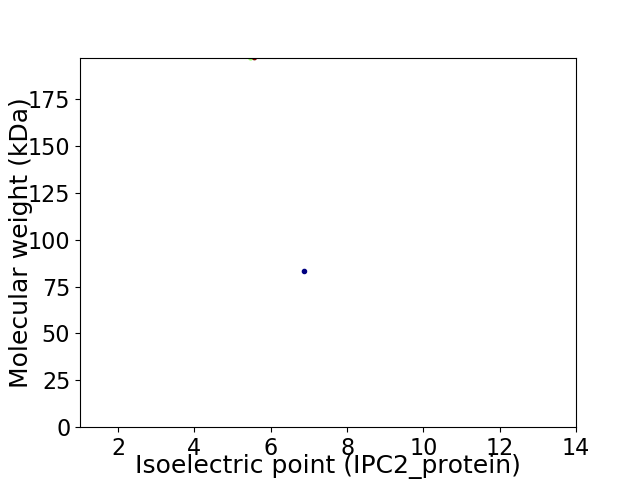
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
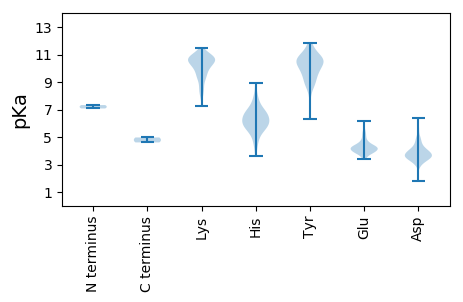
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KJ52|A0A1L3KJ52_9VIRU Uncharacterized protein OS=Shahe picorna-like virus 11 OX=1923441 PE=4 SV=1
MM1 pKa = 7.08STHH4 pKa = 5.75TQQNQTIKK12 pKa = 10.51KK13 pKa = 8.69DD14 pKa = 3.69LKK16 pKa = 10.56ILVPLFTEE24 pKa = 4.01KK25 pKa = 11.14SMDD28 pKa = 3.21IVKK31 pKa = 10.77DD32 pKa = 3.65EE33 pKa = 4.16LAKK36 pKa = 10.71ARR38 pKa = 11.84SPRR41 pKa = 11.84EE42 pKa = 3.84LDD44 pKa = 3.56VLADD48 pKa = 4.68DD49 pKa = 6.38LMTCPQCSQYY59 pKa = 11.41YY60 pKa = 9.72DD61 pKa = 3.64FCFCDD66 pKa = 4.89DD67 pKa = 4.57IYY69 pKa = 11.42DD70 pKa = 3.86SSSIPTANYY79 pKa = 9.75RR80 pKa = 11.84GVISFKK86 pKa = 9.78NVMFARR92 pKa = 11.84HH93 pKa = 6.1IKK95 pKa = 10.4HH96 pKa = 5.45KK97 pKa = 8.52TTMKK101 pKa = 10.59QMLNKK106 pKa = 9.19MRR108 pKa = 11.84VVPQSIAEE116 pKa = 4.11IFCGVNEE123 pKa = 4.17GHH125 pKa = 6.15FQSIADD131 pKa = 3.79EE132 pKa = 4.35ARR134 pKa = 11.84NGLIDD139 pKa = 3.99FRR141 pKa = 11.84SIIPKK146 pKa = 10.53LSDD149 pKa = 3.24SLNGVEE155 pKa = 6.15DD156 pKa = 4.55IISQINEE163 pKa = 3.83KK164 pKa = 9.7MPEE167 pKa = 3.52IGITIEE173 pKa = 4.33TFNEE177 pKa = 3.97TLDD180 pKa = 3.84KK181 pKa = 11.12VGSAGEE187 pKa = 4.35TINGFSDD194 pKa = 4.12KK195 pKa = 9.92GTQYY199 pKa = 11.72KK200 pKa = 10.69DD201 pKa = 3.37FALSALTIASGLYY214 pKa = 9.15WSFNVTDD221 pKa = 3.56VKK223 pKa = 10.89RR224 pKa = 11.84IGVLVCSLIALKK236 pKa = 10.66YY237 pKa = 9.89LSSKK241 pKa = 10.58EE242 pKa = 3.79FFLNMEE248 pKa = 4.38SFVGSMLDD256 pKa = 3.03RR257 pKa = 11.84VYY259 pKa = 11.05SVYY262 pKa = 10.55MNNFGSDD269 pKa = 2.97KK270 pKa = 9.78PAAKK274 pKa = 9.44IVPEE278 pKa = 4.04IGVEE282 pKa = 4.11EE283 pKa = 5.39LDD285 pKa = 3.85TLSGLLATGIAATSAIKK302 pKa = 10.64LDD304 pKa = 3.9FLSLGSVTKK313 pKa = 10.52ILADD317 pKa = 3.37STRR320 pKa = 11.84SKK322 pKa = 10.86RR323 pKa = 11.84GLEE326 pKa = 4.21DD327 pKa = 3.45CLKK330 pKa = 10.15TIVKK334 pKa = 10.05CFEE337 pKa = 3.71IVVNFVRR344 pKa = 11.84NKK346 pKa = 10.32LFSLPSMRR354 pKa = 11.84VFDD357 pKa = 4.1SSSTEE362 pKa = 3.44IDD364 pKa = 3.76DD365 pKa = 6.01FLDD368 pKa = 3.66EE369 pKa = 4.92LNEE372 pKa = 3.99FHH374 pKa = 7.62SSDD377 pKa = 4.44VNGTLPKK384 pKa = 10.31TVMTYY389 pKa = 10.72NRR391 pKa = 11.84IIDD394 pKa = 4.23FIQRR398 pKa = 11.84GKK400 pKa = 11.06VIVKK404 pKa = 9.18NIPRR408 pKa = 11.84DD409 pKa = 3.66KK410 pKa = 10.88NSEE413 pKa = 3.81GFLRR417 pKa = 11.84LVNIEE422 pKa = 4.39LKK424 pKa = 10.61YY425 pKa = 10.64LNDD428 pKa = 3.25MFEE431 pKa = 4.25VFRR434 pKa = 11.84NLNFTKK440 pKa = 10.3QGIRR444 pKa = 11.84QEE446 pKa = 4.06PVGVIFQGGAGVGKK460 pKa = 10.35SITVMHH466 pKa = 7.14LCNALSAALLSDD478 pKa = 4.42DD479 pKa = 4.52EE480 pKa = 4.63YY481 pKa = 11.63EE482 pKa = 4.16EE483 pKa = 4.69FKK485 pKa = 11.15ASASTKK491 pKa = 9.89ISVHH495 pKa = 4.43TPEE498 pKa = 3.87TKK500 pKa = 10.15FWDD503 pKa = 4.93GYY505 pKa = 10.09KK506 pKa = 9.93QGSLITVIDD515 pKa = 4.21DD516 pKa = 3.99FGQAVDD522 pKa = 3.89VPGVPDD528 pKa = 3.57NEE530 pKa = 3.98FLKK533 pKa = 10.9VIRR536 pKa = 11.84AINSYY541 pKa = 10.18EE542 pKa = 4.25YY543 pKa = 10.15IMHH546 pKa = 6.16MAKK549 pKa = 10.58LEE551 pKa = 4.32AKK553 pKa = 8.26GTTFFDD559 pKa = 3.92SRR561 pKa = 11.84FFLCTTNLVKK571 pKa = 10.78LKK573 pKa = 10.38AASIVSNEE581 pKa = 3.55ALNRR585 pKa = 11.84RR586 pKa = 11.84FPVRR590 pKa = 11.84YY591 pKa = 8.69IISIRR596 pKa = 11.84EE597 pKa = 4.08EE598 pKa = 4.33YY599 pKa = 9.63CTDD602 pKa = 3.14DD603 pKa = 3.87TVALDD608 pKa = 3.59YY609 pKa = 10.8YY610 pKa = 10.54KK611 pKa = 10.84RR612 pKa = 11.84KK613 pKa = 9.49IDD615 pKa = 3.68PTKK618 pKa = 11.0LPIKK622 pKa = 9.8QHH624 pKa = 5.77LGKK627 pKa = 10.16DD628 pKa = 3.7YY629 pKa = 11.41GSLCPQVQIFVPSDD643 pKa = 3.46DD644 pKa = 5.08DD645 pKa = 4.82GNPCGPPIGFDD656 pKa = 4.33DD657 pKa = 4.91FVQVCIHH664 pKa = 5.72EE665 pKa = 4.25HH666 pKa = 6.67DD667 pKa = 4.03KK668 pKa = 11.02RR669 pKa = 11.84KK670 pKa = 9.37NWKK673 pKa = 7.92ITQDD677 pKa = 3.44AEE679 pKa = 4.6FKK681 pKa = 10.07ATRR684 pKa = 11.84DD685 pKa = 3.17KK686 pKa = 11.45YY687 pKa = 10.61RR688 pKa = 11.84GNKK691 pKa = 8.64VEE693 pKa = 4.12EE694 pKa = 4.67PEE696 pKa = 3.97EE697 pKa = 3.86EE698 pKa = 4.73TYY700 pKa = 11.01FDD702 pKa = 4.3AEE704 pKa = 4.4NIVPEE709 pKa = 4.3SGANFSNNEE718 pKa = 3.93PYY720 pKa = 10.46DD721 pKa = 3.76VQLEE725 pKa = 4.13DD726 pKa = 3.76MEE728 pKa = 4.92EE729 pKa = 4.05KK730 pKa = 11.04LEE732 pKa = 4.17TFDD735 pKa = 5.56EE736 pKa = 4.48IVNGSKK742 pKa = 10.68PEE744 pKa = 3.87NNYY747 pKa = 9.07ICSVTSEE754 pKa = 4.11YY755 pKa = 10.72SEE757 pKa = 4.49SYY759 pKa = 10.5IRR761 pKa = 11.84VAKK764 pKa = 10.69NLVNEE769 pKa = 4.1VRR771 pKa = 11.84DD772 pKa = 3.91MKK774 pKa = 11.17VLALAKK780 pKa = 10.13EE781 pKa = 4.0KK782 pKa = 10.99FNFIFTGPLLLFHH795 pKa = 7.01LKK797 pKa = 8.57MLKK800 pKa = 10.23YY801 pKa = 10.76YY802 pKa = 8.92NQSWIKK808 pKa = 10.42AVSLGKK814 pKa = 8.43TLDD817 pKa = 3.51FLKK820 pKa = 10.99DD821 pKa = 3.32HH822 pKa = 6.55VANNPHH828 pKa = 6.66VNMLWGSRR836 pKa = 11.84RR837 pKa = 11.84YY838 pKa = 10.33FNVEE842 pKa = 3.41NAKK845 pKa = 8.64GTFQKK850 pKa = 10.75LKK852 pKa = 7.87TKK854 pKa = 9.1IWEE857 pKa = 5.05GIEE860 pKa = 3.67ILHH863 pKa = 6.05PVINYY868 pKa = 6.32VTTFAKK874 pKa = 10.35SLSWYY879 pKa = 10.49ALIFVCIATFFGLAVGAKK897 pKa = 9.72IIQLCFSLLGKK908 pKa = 10.46AYY910 pKa = 10.32DD911 pKa = 4.05FVSSAISSYY920 pKa = 10.09MGVSSVQDD928 pKa = 3.97EE929 pKa = 4.4ISSEE933 pKa = 4.29SLSKK937 pKa = 8.42TTMHH941 pKa = 6.77RR942 pKa = 11.84PKK944 pKa = 10.45GATSTYY950 pKa = 10.23IVDD953 pKa = 3.77SAKK956 pKa = 10.71AKK958 pKa = 10.75ALITPQSITDD968 pKa = 4.14LDD970 pKa = 3.72KK971 pKa = 11.41SGYY974 pKa = 10.83DD975 pKa = 3.74LMTSVLKK982 pKa = 10.81KK983 pKa = 9.2SWYY986 pKa = 8.76KK987 pKa = 10.31IEE989 pKa = 5.19MEE991 pKa = 5.07DD992 pKa = 5.43CPDD995 pKa = 3.57SNNFRR1000 pKa = 11.84LMGACFAVRR1009 pKa = 11.84GRR1011 pKa = 11.84IVLMPYY1017 pKa = 9.59HH1018 pKa = 6.57FVRR1021 pKa = 11.84KK1022 pKa = 9.53FIEE1025 pKa = 4.64GVEE1028 pKa = 4.23CVPDD1032 pKa = 3.6NQRR1035 pKa = 11.84VRR1037 pKa = 11.84VRR1039 pKa = 11.84FYY1041 pKa = 11.29KK1042 pKa = 10.65DD1043 pKa = 2.74VGGIKK1048 pKa = 10.39NSVYY1052 pKa = 9.45TYY1054 pKa = 10.81SVADD1058 pKa = 4.06IILGHH1063 pKa = 5.1VTGQLVHH1070 pKa = 6.86NDD1072 pKa = 3.24LTLVRR1077 pKa = 11.84LPEE1080 pKa = 4.28SFQVGADD1087 pKa = 3.07RR1088 pKa = 11.84VGNFAFRR1095 pKa = 11.84SDD1097 pKa = 4.07YY1098 pKa = 9.82PQHH1101 pKa = 6.69ISNIPFTMRR1110 pKa = 11.84APKK1113 pKa = 9.86TEE1115 pKa = 3.87SDD1117 pKa = 4.0VVFGEE1122 pKa = 4.4FATAVDD1128 pKa = 4.38NIVVHH1133 pKa = 6.34NSSPHH1138 pKa = 6.06CEE1140 pKa = 3.83YY1141 pKa = 10.69QIRR1144 pKa = 11.84TGYY1147 pKa = 10.1RR1148 pKa = 11.84YY1149 pKa = 10.03RR1150 pKa = 11.84GFTNGGDD1157 pKa = 3.88CGAMMVVLDD1166 pKa = 3.9NKK1168 pKa = 9.76QPKK1171 pKa = 9.47RR1172 pKa = 11.84KK1173 pKa = 9.44IFGMHH1178 pKa = 5.18VAGVSDD1184 pKa = 3.52KK1185 pKa = 10.92LVSNGYY1191 pKa = 9.48SYY1193 pKa = 11.15SASVCQEE1200 pKa = 4.55DD1201 pKa = 4.48IVADD1205 pKa = 3.98LALFGEE1211 pKa = 4.8PADD1214 pKa = 3.86NPVKK1218 pKa = 10.83EE1219 pKa = 4.46EE1220 pKa = 3.99GLILAQTDD1228 pKa = 3.35IQFGEE1233 pKa = 4.52GRR1235 pKa = 11.84FHH1237 pKa = 8.94SEE1239 pKa = 3.66GKK1241 pKa = 9.99LDD1243 pKa = 3.68KK1244 pKa = 11.07APNAVLVTSLRR1255 pKa = 11.84KK1256 pKa = 9.03SKK1258 pKa = 10.72LYY1260 pKa = 10.48GIHH1263 pKa = 6.84GEE1265 pKa = 3.94VDD1267 pKa = 3.54MEE1269 pKa = 4.45PSLLRR1274 pKa = 11.84PIEE1277 pKa = 4.42RR1278 pKa = 11.84EE1279 pKa = 3.96DD1280 pKa = 3.48GTVIDD1285 pKa = 4.46PFEE1288 pKa = 4.19VALTKK1293 pKa = 10.89YY1294 pKa = 10.41CLNRR1298 pKa = 11.84DD1299 pKa = 3.4TNINDD1304 pKa = 3.7KK1305 pKa = 10.48ALKK1308 pKa = 10.2RR1309 pKa = 11.84ATFDD1313 pKa = 4.7LEE1315 pKa = 4.13MLVMNMMPGHH1325 pKa = 6.43EE1326 pKa = 3.94PRR1328 pKa = 11.84ILNLRR1333 pKa = 11.84DD1334 pKa = 3.46VLDD1337 pKa = 4.08GTDD1340 pKa = 3.12ISGLLEE1346 pKa = 5.23GITSSTSAGYY1356 pKa = 9.78PMNLTGNVNLKK1367 pKa = 10.12KK1368 pKa = 10.37NYY1370 pKa = 8.82YY1371 pKa = 8.71SAPAGSEE1378 pKa = 3.79EE1379 pKa = 4.43RR1380 pKa = 11.84EE1381 pKa = 4.15VAFKK1385 pKa = 10.55EE1386 pKa = 3.86IEE1388 pKa = 4.03KK1389 pKa = 10.67EE1390 pKa = 3.52VDD1392 pKa = 3.23YY1393 pKa = 10.65FIAKK1397 pKa = 9.38LNSGIIPEE1405 pKa = 4.29VVYY1408 pKa = 10.71TDD1410 pKa = 3.39NLKK1413 pKa = 10.79DD1414 pKa = 3.54EE1415 pKa = 4.43RR1416 pKa = 11.84RR1417 pKa = 11.84PRR1419 pKa = 11.84AKK1421 pKa = 10.39VIAGKK1426 pKa = 8.13TRR1428 pKa = 11.84LFSGSPFIFLLVVKK1442 pKa = 9.29MYY1444 pKa = 10.88FGDD1447 pKa = 4.15FSLNYY1452 pKa = 8.78QSCRR1456 pKa = 11.84IDD1458 pKa = 3.33NEE1460 pKa = 4.16SAIGVNPYY1468 pKa = 10.34SNEE1471 pKa = 3.37WDD1473 pKa = 3.8RR1474 pKa = 11.84IARR1477 pKa = 11.84NLNVFGTGANKK1488 pKa = 10.4GAGDD1492 pKa = 3.56YY1493 pKa = 8.1TAYY1496 pKa = 10.51DD1497 pKa = 4.12GSHH1500 pKa = 7.17LPRR1503 pKa = 11.84LQLIILEE1510 pKa = 5.26LINGWYY1516 pKa = 9.28EE1517 pKa = 4.35DD1518 pKa = 4.07PQTDD1522 pKa = 4.02HH1523 pKa = 6.83IRR1525 pKa = 11.84RR1526 pKa = 11.84SLFSCITNSVHH1537 pKa = 5.97VYY1539 pKa = 10.36RR1540 pKa = 11.84GIVYY1544 pKa = 9.63DD1545 pKa = 3.7WVGGMPSGNPLTTLLNCLYY1564 pKa = 10.59NSLAFRR1570 pKa = 11.84YY1571 pKa = 8.53CWYY1574 pKa = 10.34RR1575 pKa = 11.84QDD1577 pKa = 3.39EE1578 pKa = 4.22NRR1580 pKa = 11.84PRR1582 pKa = 11.84FQTGVKK1588 pKa = 9.45LLVMGDD1594 pKa = 3.81DD1595 pKa = 4.87NIFSVHH1601 pKa = 4.83PQYY1604 pKa = 11.23RR1605 pKa = 11.84DD1606 pKa = 3.63FNEE1609 pKa = 4.13LTLPPLMEE1617 pKa = 5.37EE1618 pKa = 3.87IGLTYY1623 pKa = 10.29TPEE1626 pKa = 4.2DD1627 pKa = 3.63KK1628 pKa = 10.62EE1629 pKa = 4.28ALLEE1633 pKa = 4.15EE1634 pKa = 4.64PFRR1637 pKa = 11.84CLEE1640 pKa = 3.8QIEE1643 pKa = 4.09FLKK1646 pKa = 10.76RR1647 pKa = 11.84KK1648 pKa = 9.45FKK1650 pKa = 11.0FEE1652 pKa = 4.27PLVGAFIAPLRR1663 pKa = 11.84FSVINEE1669 pKa = 3.88MIEE1672 pKa = 3.89WTKK1675 pKa = 11.08KK1676 pKa = 8.17GTEE1679 pKa = 3.91GDD1681 pKa = 3.83KK1682 pKa = 10.05ITVEE1686 pKa = 4.14NVITALRR1693 pKa = 11.84EE1694 pKa = 3.82ASLHH1698 pKa = 5.84GKK1700 pKa = 8.06EE1701 pKa = 5.43AYY1703 pKa = 10.02DD1704 pKa = 3.6NYY1706 pKa = 10.78VSLVLPTTIGHH1717 pKa = 6.49FPGIQPSEE1725 pKa = 4.16PWIMPHH1731 pKa = 5.59SSRR1734 pKa = 11.84MFEE1737 pKa = 4.12VLNSRR1742 pKa = 11.84TFFLL1746 pKa = 4.97
MM1 pKa = 7.08STHH4 pKa = 5.75TQQNQTIKK12 pKa = 10.51KK13 pKa = 8.69DD14 pKa = 3.69LKK16 pKa = 10.56ILVPLFTEE24 pKa = 4.01KK25 pKa = 11.14SMDD28 pKa = 3.21IVKK31 pKa = 10.77DD32 pKa = 3.65EE33 pKa = 4.16LAKK36 pKa = 10.71ARR38 pKa = 11.84SPRR41 pKa = 11.84EE42 pKa = 3.84LDD44 pKa = 3.56VLADD48 pKa = 4.68DD49 pKa = 6.38LMTCPQCSQYY59 pKa = 11.41YY60 pKa = 9.72DD61 pKa = 3.64FCFCDD66 pKa = 4.89DD67 pKa = 4.57IYY69 pKa = 11.42DD70 pKa = 3.86SSSIPTANYY79 pKa = 9.75RR80 pKa = 11.84GVISFKK86 pKa = 9.78NVMFARR92 pKa = 11.84HH93 pKa = 6.1IKK95 pKa = 10.4HH96 pKa = 5.45KK97 pKa = 8.52TTMKK101 pKa = 10.59QMLNKK106 pKa = 9.19MRR108 pKa = 11.84VVPQSIAEE116 pKa = 4.11IFCGVNEE123 pKa = 4.17GHH125 pKa = 6.15FQSIADD131 pKa = 3.79EE132 pKa = 4.35ARR134 pKa = 11.84NGLIDD139 pKa = 3.99FRR141 pKa = 11.84SIIPKK146 pKa = 10.53LSDD149 pKa = 3.24SLNGVEE155 pKa = 6.15DD156 pKa = 4.55IISQINEE163 pKa = 3.83KK164 pKa = 9.7MPEE167 pKa = 3.52IGITIEE173 pKa = 4.33TFNEE177 pKa = 3.97TLDD180 pKa = 3.84KK181 pKa = 11.12VGSAGEE187 pKa = 4.35TINGFSDD194 pKa = 4.12KK195 pKa = 9.92GTQYY199 pKa = 11.72KK200 pKa = 10.69DD201 pKa = 3.37FALSALTIASGLYY214 pKa = 9.15WSFNVTDD221 pKa = 3.56VKK223 pKa = 10.89RR224 pKa = 11.84IGVLVCSLIALKK236 pKa = 10.66YY237 pKa = 9.89LSSKK241 pKa = 10.58EE242 pKa = 3.79FFLNMEE248 pKa = 4.38SFVGSMLDD256 pKa = 3.03RR257 pKa = 11.84VYY259 pKa = 11.05SVYY262 pKa = 10.55MNNFGSDD269 pKa = 2.97KK270 pKa = 9.78PAAKK274 pKa = 9.44IVPEE278 pKa = 4.04IGVEE282 pKa = 4.11EE283 pKa = 5.39LDD285 pKa = 3.85TLSGLLATGIAATSAIKK302 pKa = 10.64LDD304 pKa = 3.9FLSLGSVTKK313 pKa = 10.52ILADD317 pKa = 3.37STRR320 pKa = 11.84SKK322 pKa = 10.86RR323 pKa = 11.84GLEE326 pKa = 4.21DD327 pKa = 3.45CLKK330 pKa = 10.15TIVKK334 pKa = 10.05CFEE337 pKa = 3.71IVVNFVRR344 pKa = 11.84NKK346 pKa = 10.32LFSLPSMRR354 pKa = 11.84VFDD357 pKa = 4.1SSSTEE362 pKa = 3.44IDD364 pKa = 3.76DD365 pKa = 6.01FLDD368 pKa = 3.66EE369 pKa = 4.92LNEE372 pKa = 3.99FHH374 pKa = 7.62SSDD377 pKa = 4.44VNGTLPKK384 pKa = 10.31TVMTYY389 pKa = 10.72NRR391 pKa = 11.84IIDD394 pKa = 4.23FIQRR398 pKa = 11.84GKK400 pKa = 11.06VIVKK404 pKa = 9.18NIPRR408 pKa = 11.84DD409 pKa = 3.66KK410 pKa = 10.88NSEE413 pKa = 3.81GFLRR417 pKa = 11.84LVNIEE422 pKa = 4.39LKK424 pKa = 10.61YY425 pKa = 10.64LNDD428 pKa = 3.25MFEE431 pKa = 4.25VFRR434 pKa = 11.84NLNFTKK440 pKa = 10.3QGIRR444 pKa = 11.84QEE446 pKa = 4.06PVGVIFQGGAGVGKK460 pKa = 10.35SITVMHH466 pKa = 7.14LCNALSAALLSDD478 pKa = 4.42DD479 pKa = 4.52EE480 pKa = 4.63YY481 pKa = 11.63EE482 pKa = 4.16EE483 pKa = 4.69FKK485 pKa = 11.15ASASTKK491 pKa = 9.89ISVHH495 pKa = 4.43TPEE498 pKa = 3.87TKK500 pKa = 10.15FWDD503 pKa = 4.93GYY505 pKa = 10.09KK506 pKa = 9.93QGSLITVIDD515 pKa = 4.21DD516 pKa = 3.99FGQAVDD522 pKa = 3.89VPGVPDD528 pKa = 3.57NEE530 pKa = 3.98FLKK533 pKa = 10.9VIRR536 pKa = 11.84AINSYY541 pKa = 10.18EE542 pKa = 4.25YY543 pKa = 10.15IMHH546 pKa = 6.16MAKK549 pKa = 10.58LEE551 pKa = 4.32AKK553 pKa = 8.26GTTFFDD559 pKa = 3.92SRR561 pKa = 11.84FFLCTTNLVKK571 pKa = 10.78LKK573 pKa = 10.38AASIVSNEE581 pKa = 3.55ALNRR585 pKa = 11.84RR586 pKa = 11.84FPVRR590 pKa = 11.84YY591 pKa = 8.69IISIRR596 pKa = 11.84EE597 pKa = 4.08EE598 pKa = 4.33YY599 pKa = 9.63CTDD602 pKa = 3.14DD603 pKa = 3.87TVALDD608 pKa = 3.59YY609 pKa = 10.8YY610 pKa = 10.54KK611 pKa = 10.84RR612 pKa = 11.84KK613 pKa = 9.49IDD615 pKa = 3.68PTKK618 pKa = 11.0LPIKK622 pKa = 9.8QHH624 pKa = 5.77LGKK627 pKa = 10.16DD628 pKa = 3.7YY629 pKa = 11.41GSLCPQVQIFVPSDD643 pKa = 3.46DD644 pKa = 5.08DD645 pKa = 4.82GNPCGPPIGFDD656 pKa = 4.33DD657 pKa = 4.91FVQVCIHH664 pKa = 5.72EE665 pKa = 4.25HH666 pKa = 6.67DD667 pKa = 4.03KK668 pKa = 11.02RR669 pKa = 11.84KK670 pKa = 9.37NWKK673 pKa = 7.92ITQDD677 pKa = 3.44AEE679 pKa = 4.6FKK681 pKa = 10.07ATRR684 pKa = 11.84DD685 pKa = 3.17KK686 pKa = 11.45YY687 pKa = 10.61RR688 pKa = 11.84GNKK691 pKa = 8.64VEE693 pKa = 4.12EE694 pKa = 4.67PEE696 pKa = 3.97EE697 pKa = 3.86EE698 pKa = 4.73TYY700 pKa = 11.01FDD702 pKa = 4.3AEE704 pKa = 4.4NIVPEE709 pKa = 4.3SGANFSNNEE718 pKa = 3.93PYY720 pKa = 10.46DD721 pKa = 3.76VQLEE725 pKa = 4.13DD726 pKa = 3.76MEE728 pKa = 4.92EE729 pKa = 4.05KK730 pKa = 11.04LEE732 pKa = 4.17TFDD735 pKa = 5.56EE736 pKa = 4.48IVNGSKK742 pKa = 10.68PEE744 pKa = 3.87NNYY747 pKa = 9.07ICSVTSEE754 pKa = 4.11YY755 pKa = 10.72SEE757 pKa = 4.49SYY759 pKa = 10.5IRR761 pKa = 11.84VAKK764 pKa = 10.69NLVNEE769 pKa = 4.1VRR771 pKa = 11.84DD772 pKa = 3.91MKK774 pKa = 11.17VLALAKK780 pKa = 10.13EE781 pKa = 4.0KK782 pKa = 10.99FNFIFTGPLLLFHH795 pKa = 7.01LKK797 pKa = 8.57MLKK800 pKa = 10.23YY801 pKa = 10.76YY802 pKa = 8.92NQSWIKK808 pKa = 10.42AVSLGKK814 pKa = 8.43TLDD817 pKa = 3.51FLKK820 pKa = 10.99DD821 pKa = 3.32HH822 pKa = 6.55VANNPHH828 pKa = 6.66VNMLWGSRR836 pKa = 11.84RR837 pKa = 11.84YY838 pKa = 10.33FNVEE842 pKa = 3.41NAKK845 pKa = 8.64GTFQKK850 pKa = 10.75LKK852 pKa = 7.87TKK854 pKa = 9.1IWEE857 pKa = 5.05GIEE860 pKa = 3.67ILHH863 pKa = 6.05PVINYY868 pKa = 6.32VTTFAKK874 pKa = 10.35SLSWYY879 pKa = 10.49ALIFVCIATFFGLAVGAKK897 pKa = 9.72IIQLCFSLLGKK908 pKa = 10.46AYY910 pKa = 10.32DD911 pKa = 4.05FVSSAISSYY920 pKa = 10.09MGVSSVQDD928 pKa = 3.97EE929 pKa = 4.4ISSEE933 pKa = 4.29SLSKK937 pKa = 8.42TTMHH941 pKa = 6.77RR942 pKa = 11.84PKK944 pKa = 10.45GATSTYY950 pKa = 10.23IVDD953 pKa = 3.77SAKK956 pKa = 10.71AKK958 pKa = 10.75ALITPQSITDD968 pKa = 4.14LDD970 pKa = 3.72KK971 pKa = 11.41SGYY974 pKa = 10.83DD975 pKa = 3.74LMTSVLKK982 pKa = 10.81KK983 pKa = 9.2SWYY986 pKa = 8.76KK987 pKa = 10.31IEE989 pKa = 5.19MEE991 pKa = 5.07DD992 pKa = 5.43CPDD995 pKa = 3.57SNNFRR1000 pKa = 11.84LMGACFAVRR1009 pKa = 11.84GRR1011 pKa = 11.84IVLMPYY1017 pKa = 9.59HH1018 pKa = 6.57FVRR1021 pKa = 11.84KK1022 pKa = 9.53FIEE1025 pKa = 4.64GVEE1028 pKa = 4.23CVPDD1032 pKa = 3.6NQRR1035 pKa = 11.84VRR1037 pKa = 11.84VRR1039 pKa = 11.84FYY1041 pKa = 11.29KK1042 pKa = 10.65DD1043 pKa = 2.74VGGIKK1048 pKa = 10.39NSVYY1052 pKa = 9.45TYY1054 pKa = 10.81SVADD1058 pKa = 4.06IILGHH1063 pKa = 5.1VTGQLVHH1070 pKa = 6.86NDD1072 pKa = 3.24LTLVRR1077 pKa = 11.84LPEE1080 pKa = 4.28SFQVGADD1087 pKa = 3.07RR1088 pKa = 11.84VGNFAFRR1095 pKa = 11.84SDD1097 pKa = 4.07YY1098 pKa = 9.82PQHH1101 pKa = 6.69ISNIPFTMRR1110 pKa = 11.84APKK1113 pKa = 9.86TEE1115 pKa = 3.87SDD1117 pKa = 4.0VVFGEE1122 pKa = 4.4FATAVDD1128 pKa = 4.38NIVVHH1133 pKa = 6.34NSSPHH1138 pKa = 6.06CEE1140 pKa = 3.83YY1141 pKa = 10.69QIRR1144 pKa = 11.84TGYY1147 pKa = 10.1RR1148 pKa = 11.84YY1149 pKa = 10.03RR1150 pKa = 11.84GFTNGGDD1157 pKa = 3.88CGAMMVVLDD1166 pKa = 3.9NKK1168 pKa = 9.76QPKK1171 pKa = 9.47RR1172 pKa = 11.84KK1173 pKa = 9.44IFGMHH1178 pKa = 5.18VAGVSDD1184 pKa = 3.52KK1185 pKa = 10.92LVSNGYY1191 pKa = 9.48SYY1193 pKa = 11.15SASVCQEE1200 pKa = 4.55DD1201 pKa = 4.48IVADD1205 pKa = 3.98LALFGEE1211 pKa = 4.8PADD1214 pKa = 3.86NPVKK1218 pKa = 10.83EE1219 pKa = 4.46EE1220 pKa = 3.99GLILAQTDD1228 pKa = 3.35IQFGEE1233 pKa = 4.52GRR1235 pKa = 11.84FHH1237 pKa = 8.94SEE1239 pKa = 3.66GKK1241 pKa = 9.99LDD1243 pKa = 3.68KK1244 pKa = 11.07APNAVLVTSLRR1255 pKa = 11.84KK1256 pKa = 9.03SKK1258 pKa = 10.72LYY1260 pKa = 10.48GIHH1263 pKa = 6.84GEE1265 pKa = 3.94VDD1267 pKa = 3.54MEE1269 pKa = 4.45PSLLRR1274 pKa = 11.84PIEE1277 pKa = 4.42RR1278 pKa = 11.84EE1279 pKa = 3.96DD1280 pKa = 3.48GTVIDD1285 pKa = 4.46PFEE1288 pKa = 4.19VALTKK1293 pKa = 10.89YY1294 pKa = 10.41CLNRR1298 pKa = 11.84DD1299 pKa = 3.4TNINDD1304 pKa = 3.7KK1305 pKa = 10.48ALKK1308 pKa = 10.2RR1309 pKa = 11.84ATFDD1313 pKa = 4.7LEE1315 pKa = 4.13MLVMNMMPGHH1325 pKa = 6.43EE1326 pKa = 3.94PRR1328 pKa = 11.84ILNLRR1333 pKa = 11.84DD1334 pKa = 3.46VLDD1337 pKa = 4.08GTDD1340 pKa = 3.12ISGLLEE1346 pKa = 5.23GITSSTSAGYY1356 pKa = 9.78PMNLTGNVNLKK1367 pKa = 10.12KK1368 pKa = 10.37NYY1370 pKa = 8.82YY1371 pKa = 8.71SAPAGSEE1378 pKa = 3.79EE1379 pKa = 4.43RR1380 pKa = 11.84EE1381 pKa = 4.15VAFKK1385 pKa = 10.55EE1386 pKa = 3.86IEE1388 pKa = 4.03KK1389 pKa = 10.67EE1390 pKa = 3.52VDD1392 pKa = 3.23YY1393 pKa = 10.65FIAKK1397 pKa = 9.38LNSGIIPEE1405 pKa = 4.29VVYY1408 pKa = 10.71TDD1410 pKa = 3.39NLKK1413 pKa = 10.79DD1414 pKa = 3.54EE1415 pKa = 4.43RR1416 pKa = 11.84RR1417 pKa = 11.84PRR1419 pKa = 11.84AKK1421 pKa = 10.39VIAGKK1426 pKa = 8.13TRR1428 pKa = 11.84LFSGSPFIFLLVVKK1442 pKa = 9.29MYY1444 pKa = 10.88FGDD1447 pKa = 4.15FSLNYY1452 pKa = 8.78QSCRR1456 pKa = 11.84IDD1458 pKa = 3.33NEE1460 pKa = 4.16SAIGVNPYY1468 pKa = 10.34SNEE1471 pKa = 3.37WDD1473 pKa = 3.8RR1474 pKa = 11.84IARR1477 pKa = 11.84NLNVFGTGANKK1488 pKa = 10.4GAGDD1492 pKa = 3.56YY1493 pKa = 8.1TAYY1496 pKa = 10.51DD1497 pKa = 4.12GSHH1500 pKa = 7.17LPRR1503 pKa = 11.84LQLIILEE1510 pKa = 5.26LINGWYY1516 pKa = 9.28EE1517 pKa = 4.35DD1518 pKa = 4.07PQTDD1522 pKa = 4.02HH1523 pKa = 6.83IRR1525 pKa = 11.84RR1526 pKa = 11.84SLFSCITNSVHH1537 pKa = 5.97VYY1539 pKa = 10.36RR1540 pKa = 11.84GIVYY1544 pKa = 9.63DD1545 pKa = 3.7WVGGMPSGNPLTTLLNCLYY1564 pKa = 10.59NSLAFRR1570 pKa = 11.84YY1571 pKa = 8.53CWYY1574 pKa = 10.34RR1575 pKa = 11.84QDD1577 pKa = 3.39EE1578 pKa = 4.22NRR1580 pKa = 11.84PRR1582 pKa = 11.84FQTGVKK1588 pKa = 9.45LLVMGDD1594 pKa = 3.81DD1595 pKa = 4.87NIFSVHH1601 pKa = 4.83PQYY1604 pKa = 11.23RR1605 pKa = 11.84DD1606 pKa = 3.63FNEE1609 pKa = 4.13LTLPPLMEE1617 pKa = 5.37EE1618 pKa = 3.87IGLTYY1623 pKa = 10.29TPEE1626 pKa = 4.2DD1627 pKa = 3.63KK1628 pKa = 10.62EE1629 pKa = 4.28ALLEE1633 pKa = 4.15EE1634 pKa = 4.64PFRR1637 pKa = 11.84CLEE1640 pKa = 3.8QIEE1643 pKa = 4.09FLKK1646 pKa = 10.76RR1647 pKa = 11.84KK1648 pKa = 9.45FKK1650 pKa = 11.0FEE1652 pKa = 4.27PLVGAFIAPLRR1663 pKa = 11.84FSVINEE1669 pKa = 3.88MIEE1672 pKa = 3.89WTKK1675 pKa = 11.08KK1676 pKa = 8.17GTEE1679 pKa = 3.91GDD1681 pKa = 3.83KK1682 pKa = 10.05ITVEE1686 pKa = 4.14NVITALRR1693 pKa = 11.84EE1694 pKa = 3.82ASLHH1698 pKa = 5.84GKK1700 pKa = 8.06EE1701 pKa = 5.43AYY1703 pKa = 10.02DD1704 pKa = 3.6NYY1706 pKa = 10.78VSLVLPTTIGHH1717 pKa = 6.49FPGIQPSEE1725 pKa = 4.16PWIMPHH1731 pKa = 5.59SSRR1734 pKa = 11.84MFEE1737 pKa = 4.12VLNSRR1742 pKa = 11.84TFFLL1746 pKa = 4.97
Molecular weight: 197.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KJ52|A0A1L3KJ52_9VIRU Uncharacterized protein OS=Shahe picorna-like virus 11 OX=1923441 PE=4 SV=1
MM1 pKa = 7.32FSSSSDD7 pKa = 3.41NFNQDD12 pKa = 1.8IKK14 pKa = 10.24TFLSKK19 pKa = 10.49PVILQSNTLGNTDD32 pKa = 3.71TVSSFPTMLMPSTLIQTFSTINNKK56 pKa = 9.3LQGYY60 pKa = 8.89LGIRR64 pKa = 11.84ATMVFRR70 pKa = 11.84LQVNANPFQQGRR82 pKa = 11.84YY83 pKa = 6.5MLTWVPLGGTGINAATVGTHH103 pKa = 5.81YY104 pKa = 10.74NPHH107 pKa = 5.94IQSLVQRR114 pKa = 11.84STLPRR119 pKa = 11.84VEE121 pKa = 5.59LDD123 pKa = 3.74LACDD127 pKa = 3.27THH129 pKa = 7.83VVLRR133 pKa = 11.84VPYY136 pKa = 9.31VSSKK140 pKa = 10.81NFYY143 pKa = 9.48PLAGQSSTEE152 pKa = 3.9LFGSLGYY159 pKa = 8.79LTLFPYY165 pKa = 9.14VTLNGIVNQTAGYY178 pKa = 6.95TIWGHH183 pKa = 5.96FEE185 pKa = 4.99DD186 pKa = 4.63IEE188 pKa = 4.45LVGVTVPQSGAGFSNTDD205 pKa = 3.37RR206 pKa = 11.84EE207 pKa = 4.33AKK209 pKa = 10.49AIGVQPLSNSLSLISKK225 pKa = 10.65ASGTLSKK232 pKa = 11.18VPFIGDD238 pKa = 3.58YY239 pKa = 9.89ATGISWLSEE248 pKa = 3.89RR249 pKa = 11.84LAKK252 pKa = 9.52TAMVFGWSRR261 pKa = 11.84PASMEE266 pKa = 3.58PTKK269 pKa = 10.83RR270 pKa = 11.84MYY272 pKa = 10.67RR273 pKa = 11.84STLQDD278 pKa = 3.48YY279 pKa = 10.88CNVDD283 pKa = 3.32AVDD286 pKa = 3.66YY287 pKa = 11.04SQMLSLSSKK296 pKa = 9.76NQLHH300 pKa = 5.65VLPGFSGTNTDD311 pKa = 3.48EE312 pKa = 4.21MDD314 pKa = 3.59FSFIASIPAFDD325 pKa = 6.05QLITWNFSDD334 pKa = 5.57GIGLRR339 pKa = 11.84KK340 pKa = 7.23MTKK343 pKa = 8.81EE344 pKa = 3.78VCPLEE349 pKa = 4.15NVNNFTLAFGGVVANNYY366 pKa = 9.85KK367 pKa = 10.58PIDD370 pKa = 3.58FVSRR374 pKa = 11.84HH375 pKa = 3.62FTYY378 pKa = 9.4WRR380 pKa = 11.84GSIVYY385 pKa = 7.76TFKK388 pKa = 10.55FVKK391 pKa = 10.04TNFHH395 pKa = 6.32SGRR398 pKa = 11.84LAICFAPFEE407 pKa = 5.65DD408 pKa = 4.22ISSAPRR414 pKa = 11.84TLTLPLSAFTHH425 pKa = 5.39RR426 pKa = 11.84HH427 pKa = 4.49IVDD430 pKa = 2.92IRR432 pKa = 11.84EE433 pKa = 4.24TNMVTLTIPYY443 pKa = 9.69ISSTPFRR450 pKa = 11.84NTLVGNGNSTGLIEE464 pKa = 5.18IYY466 pKa = 10.74VIDD469 pKa = 4.95PLTGPSTVPNQIPIIVEE486 pKa = 3.95ISAGPDD492 pKa = 3.11FEE494 pKa = 4.8VAVPRR499 pKa = 11.84QLNHH503 pKa = 6.48APAVNVVPQSGAGFSNTNSACAMTDD528 pKa = 2.92TTLGVSSMRR537 pKa = 11.84SDD539 pKa = 3.53DD540 pKa = 4.09HH541 pKa = 8.07LNAAACVGEE550 pKa = 4.71KK551 pKa = 10.0ISSFRR556 pKa = 11.84TMLKK560 pKa = 10.06SFNFLPYY567 pKa = 10.24QNPAAAANKK576 pKa = 9.08FLNIAPFNTPVYY588 pKa = 10.11FDD590 pKa = 4.1AVANAASTQVADD602 pKa = 5.01LYY604 pKa = 10.7TILSSIYY611 pKa = 9.08LFSRR615 pKa = 11.84GSVRR619 pKa = 11.84LKK621 pKa = 10.72INMTGASGASSSNSFSASLFAVPVDD646 pKa = 3.57NLSISSISNLNALGLTGTTAVSTSNILRR674 pKa = 11.84TYY676 pKa = 10.58SHH678 pKa = 7.48LGQNLCAEE686 pKa = 4.33IQVPQYY692 pKa = 11.83NMMHH696 pKa = 6.06SRR698 pKa = 11.84VNWEE702 pKa = 4.23HH703 pKa = 5.71VASPNISTPVFSGRR717 pKa = 11.84SKK719 pKa = 10.78VVVSITNLSMNIEE732 pKa = 4.18PTVGIHH738 pKa = 6.72RR739 pKa = 11.84AGGDD743 pKa = 3.5DD744 pKa = 4.01CNFGCFISIPPMVSVMAGTVPII766 pKa = 4.61
MM1 pKa = 7.32FSSSSDD7 pKa = 3.41NFNQDD12 pKa = 1.8IKK14 pKa = 10.24TFLSKK19 pKa = 10.49PVILQSNTLGNTDD32 pKa = 3.71TVSSFPTMLMPSTLIQTFSTINNKK56 pKa = 9.3LQGYY60 pKa = 8.89LGIRR64 pKa = 11.84ATMVFRR70 pKa = 11.84LQVNANPFQQGRR82 pKa = 11.84YY83 pKa = 6.5MLTWVPLGGTGINAATVGTHH103 pKa = 5.81YY104 pKa = 10.74NPHH107 pKa = 5.94IQSLVQRR114 pKa = 11.84STLPRR119 pKa = 11.84VEE121 pKa = 5.59LDD123 pKa = 3.74LACDD127 pKa = 3.27THH129 pKa = 7.83VVLRR133 pKa = 11.84VPYY136 pKa = 9.31VSSKK140 pKa = 10.81NFYY143 pKa = 9.48PLAGQSSTEE152 pKa = 3.9LFGSLGYY159 pKa = 8.79LTLFPYY165 pKa = 9.14VTLNGIVNQTAGYY178 pKa = 6.95TIWGHH183 pKa = 5.96FEE185 pKa = 4.99DD186 pKa = 4.63IEE188 pKa = 4.45LVGVTVPQSGAGFSNTDD205 pKa = 3.37RR206 pKa = 11.84EE207 pKa = 4.33AKK209 pKa = 10.49AIGVQPLSNSLSLISKK225 pKa = 10.65ASGTLSKK232 pKa = 11.18VPFIGDD238 pKa = 3.58YY239 pKa = 9.89ATGISWLSEE248 pKa = 3.89RR249 pKa = 11.84LAKK252 pKa = 9.52TAMVFGWSRR261 pKa = 11.84PASMEE266 pKa = 3.58PTKK269 pKa = 10.83RR270 pKa = 11.84MYY272 pKa = 10.67RR273 pKa = 11.84STLQDD278 pKa = 3.48YY279 pKa = 10.88CNVDD283 pKa = 3.32AVDD286 pKa = 3.66YY287 pKa = 11.04SQMLSLSSKK296 pKa = 9.76NQLHH300 pKa = 5.65VLPGFSGTNTDD311 pKa = 3.48EE312 pKa = 4.21MDD314 pKa = 3.59FSFIASIPAFDD325 pKa = 6.05QLITWNFSDD334 pKa = 5.57GIGLRR339 pKa = 11.84KK340 pKa = 7.23MTKK343 pKa = 8.81EE344 pKa = 3.78VCPLEE349 pKa = 4.15NVNNFTLAFGGVVANNYY366 pKa = 9.85KK367 pKa = 10.58PIDD370 pKa = 3.58FVSRR374 pKa = 11.84HH375 pKa = 3.62FTYY378 pKa = 9.4WRR380 pKa = 11.84GSIVYY385 pKa = 7.76TFKK388 pKa = 10.55FVKK391 pKa = 10.04TNFHH395 pKa = 6.32SGRR398 pKa = 11.84LAICFAPFEE407 pKa = 5.65DD408 pKa = 4.22ISSAPRR414 pKa = 11.84TLTLPLSAFTHH425 pKa = 5.39RR426 pKa = 11.84HH427 pKa = 4.49IVDD430 pKa = 2.92IRR432 pKa = 11.84EE433 pKa = 4.24TNMVTLTIPYY443 pKa = 9.69ISSTPFRR450 pKa = 11.84NTLVGNGNSTGLIEE464 pKa = 5.18IYY466 pKa = 10.74VIDD469 pKa = 4.95PLTGPSTVPNQIPIIVEE486 pKa = 3.95ISAGPDD492 pKa = 3.11FEE494 pKa = 4.8VAVPRR499 pKa = 11.84QLNHH503 pKa = 6.48APAVNVVPQSGAGFSNTNSACAMTDD528 pKa = 2.92TTLGVSSMRR537 pKa = 11.84SDD539 pKa = 3.53DD540 pKa = 4.09HH541 pKa = 8.07LNAAACVGEE550 pKa = 4.71KK551 pKa = 10.0ISSFRR556 pKa = 11.84TMLKK560 pKa = 10.06SFNFLPYY567 pKa = 10.24QNPAAAANKK576 pKa = 9.08FLNIAPFNTPVYY588 pKa = 10.11FDD590 pKa = 4.1AVANAASTQVADD602 pKa = 5.01LYY604 pKa = 10.7TILSSIYY611 pKa = 9.08LFSRR615 pKa = 11.84GSVRR619 pKa = 11.84LKK621 pKa = 10.72INMTGASGASSSNSFSASLFAVPVDD646 pKa = 3.57NLSISSISNLNALGLTGTTAVSTSNILRR674 pKa = 11.84TYY676 pKa = 10.58SHH678 pKa = 7.48LGQNLCAEE686 pKa = 4.33IQVPQYY692 pKa = 11.83NMMHH696 pKa = 6.06SRR698 pKa = 11.84VNWEE702 pKa = 4.23HH703 pKa = 5.71VASPNISTPVFSGRR717 pKa = 11.84SKK719 pKa = 10.78VVVSITNLSMNIEE732 pKa = 4.18PTVGIHH738 pKa = 6.72RR739 pKa = 11.84AGGDD743 pKa = 3.5DD744 pKa = 4.01CNFGCFISIPPMVSVMAGTVPII766 pKa = 4.61
Molecular weight: 83.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2512 |
766 |
1746 |
1256.0 |
140.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.892 ± 0.643 | 1.513 ± 0.188 |
5.533 ± 1.043 | 5.295 ± 1.563 |
5.693 ± 0.029 | 6.369 ± 0.16 |
1.911 ± 0.026 | 6.807 ± 0.228 |
5.374 ± 1.535 | 8.519 ± 0.091 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.588 ± 0.086 | 5.932 ± 0.621 |
4.658 ± 0.604 | 2.747 ± 0.287 |
4.299 ± 0.358 | 8.519 ± 1.504 |
6.29 ± 1.147 | 7.604 ± 0.2 |
0.836 ± 0.043 | 3.623 ± 0.344 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
