
Flavobacterium phage vB_FspP_elemoA_3-5A
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Podoviridae; unclassified Podoviridae; crAss-like viruses
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
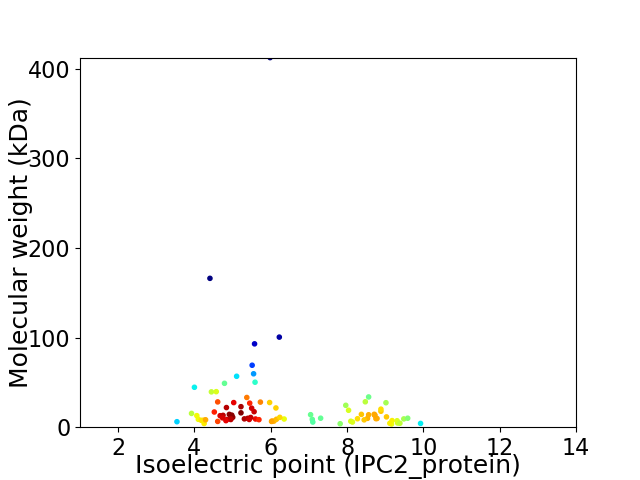
Virtual 2D-PAGE plot for 90 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A7D7JNV7|A0A7D7JNV7_9CAUD Uncharacterized protein OS=Flavobacterium phage vB_FspP_elemoA_3-5A OX=2743819 GN=elemo35A_phanotate89 PE=4 SV=1
MM1 pKa = 7.43ARR3 pKa = 11.84IKK5 pKa = 10.03TYY7 pKa = 10.42TIDD10 pKa = 3.75TLISDD15 pKa = 3.92NDD17 pKa = 3.88IIIGSDD23 pKa = 2.83ADD25 pKa = 3.98NNNEE29 pKa = 4.18TKK31 pKa = 10.68NFSAGLIRR39 pKa = 11.84EE40 pKa = 4.38FVLSGLEE47 pKa = 4.02PEE49 pKa = 4.48VGGNLKK55 pKa = 8.7ITTIVDD61 pKa = 3.24NDD63 pKa = 3.97SEE65 pKa = 4.56EE66 pKa = 4.43TTPEE70 pKa = 3.99DD71 pKa = 3.59YY72 pKa = 10.94FNNSVTPIIVLHH84 pKa = 5.26YY85 pKa = 9.83EE86 pKa = 3.83IVFLILNGRR95 pKa = 11.84TFIFRR100 pKa = 11.84KK101 pKa = 10.51NNDD104 pKa = 3.08TYY106 pKa = 11.57GVDD109 pKa = 3.37EE110 pKa = 4.44TQVVSGDD117 pKa = 3.4FTEE120 pKa = 4.8IDD122 pKa = 3.08ITSVINANLQDD133 pKa = 4.38LDD135 pKa = 4.02SVLTEE140 pKa = 4.25GNEE143 pKa = 4.8APDD146 pKa = 3.64KK147 pKa = 10.42DD148 pKa = 3.84AKK150 pKa = 10.74IRR152 pKa = 11.84DD153 pKa = 3.64LYY155 pKa = 11.22LYY157 pKa = 10.55DD158 pKa = 5.55DD159 pKa = 4.97FGSPDD164 pKa = 3.27GYY166 pKa = 11.61ARR168 pKa = 11.84LYY170 pKa = 10.82SDD172 pKa = 3.34KK173 pKa = 10.93QSLYY177 pKa = 10.94FMNKK181 pKa = 7.41TGEE184 pKa = 4.39SVFQLEE190 pKa = 4.51KK191 pKa = 10.67NSLYY195 pKa = 10.65FPIGAYY201 pKa = 7.89TYY203 pKa = 10.67KK204 pKa = 9.39ITTPSITGGRR214 pKa = 11.84IATFQDD220 pKa = 2.86ASGIVAYY227 pKa = 9.58TSDD230 pKa = 3.7IPTDD234 pKa = 4.03YY235 pKa = 10.41ISSITSSSDD244 pKa = 2.96EE245 pKa = 4.02LDD247 pKa = 3.08ITTVAGEE254 pKa = 4.03TTIIYY259 pKa = 8.06TPKK262 pKa = 10.58KK263 pKa = 9.09EE264 pKa = 3.77IQLIGTEE271 pKa = 4.25LSNNLTNTTFFSNGGEE287 pKa = 3.9FSYY290 pKa = 11.47DD291 pKa = 2.99EE292 pKa = 4.13TLYY295 pKa = 10.5PITALDD301 pKa = 3.86LKK303 pKa = 10.84FNYY306 pKa = 10.62NIDD309 pKa = 3.72DD310 pKa = 3.73NGIDD314 pKa = 3.25FGYY317 pKa = 11.01YY318 pKa = 8.69VFHH321 pKa = 7.66YY322 pKa = 10.39EE323 pKa = 4.68DD324 pKa = 3.95GRR326 pKa = 11.84NVIAPIEE333 pKa = 3.91LAGSIISGTEE343 pKa = 3.29LRR345 pKa = 11.84VRR347 pKa = 11.84LDD349 pKa = 3.19LATYY353 pKa = 6.96WTEE356 pKa = 4.13SPDD359 pKa = 3.24PRR361 pKa = 11.84LSYY364 pKa = 10.93FEE366 pKa = 4.47INLYY370 pKa = 10.15QGTLISEE377 pKa = 4.53DD378 pKa = 3.04GVYY381 pKa = 10.6FDD383 pKa = 4.44GHH385 pKa = 6.54APSSQIKK392 pKa = 9.75YY393 pKa = 9.9FNIWTT398 pKa = 3.82
MM1 pKa = 7.43ARR3 pKa = 11.84IKK5 pKa = 10.03TYY7 pKa = 10.42TIDD10 pKa = 3.75TLISDD15 pKa = 3.92NDD17 pKa = 3.88IIIGSDD23 pKa = 2.83ADD25 pKa = 3.98NNNEE29 pKa = 4.18TKK31 pKa = 10.68NFSAGLIRR39 pKa = 11.84EE40 pKa = 4.38FVLSGLEE47 pKa = 4.02PEE49 pKa = 4.48VGGNLKK55 pKa = 8.7ITTIVDD61 pKa = 3.24NDD63 pKa = 3.97SEE65 pKa = 4.56EE66 pKa = 4.43TTPEE70 pKa = 3.99DD71 pKa = 3.59YY72 pKa = 10.94FNNSVTPIIVLHH84 pKa = 5.26YY85 pKa = 9.83EE86 pKa = 3.83IVFLILNGRR95 pKa = 11.84TFIFRR100 pKa = 11.84KK101 pKa = 10.51NNDD104 pKa = 3.08TYY106 pKa = 11.57GVDD109 pKa = 3.37EE110 pKa = 4.44TQVVSGDD117 pKa = 3.4FTEE120 pKa = 4.8IDD122 pKa = 3.08ITSVINANLQDD133 pKa = 4.38LDD135 pKa = 4.02SVLTEE140 pKa = 4.25GNEE143 pKa = 4.8APDD146 pKa = 3.64KK147 pKa = 10.42DD148 pKa = 3.84AKK150 pKa = 10.74IRR152 pKa = 11.84DD153 pKa = 3.64LYY155 pKa = 11.22LYY157 pKa = 10.55DD158 pKa = 5.55DD159 pKa = 4.97FGSPDD164 pKa = 3.27GYY166 pKa = 11.61ARR168 pKa = 11.84LYY170 pKa = 10.82SDD172 pKa = 3.34KK173 pKa = 10.93QSLYY177 pKa = 10.94FMNKK181 pKa = 7.41TGEE184 pKa = 4.39SVFQLEE190 pKa = 4.51KK191 pKa = 10.67NSLYY195 pKa = 10.65FPIGAYY201 pKa = 7.89TYY203 pKa = 10.67KK204 pKa = 9.39ITTPSITGGRR214 pKa = 11.84IATFQDD220 pKa = 2.86ASGIVAYY227 pKa = 9.58TSDD230 pKa = 3.7IPTDD234 pKa = 4.03YY235 pKa = 10.41ISSITSSSDD244 pKa = 2.96EE245 pKa = 4.02LDD247 pKa = 3.08ITTVAGEE254 pKa = 4.03TTIIYY259 pKa = 8.06TPKK262 pKa = 10.58KK263 pKa = 9.09EE264 pKa = 3.77IQLIGTEE271 pKa = 4.25LSNNLTNTTFFSNGGEE287 pKa = 3.9FSYY290 pKa = 11.47DD291 pKa = 2.99EE292 pKa = 4.13TLYY295 pKa = 10.5PITALDD301 pKa = 3.86LKK303 pKa = 10.84FNYY306 pKa = 10.62NIDD309 pKa = 3.72DD310 pKa = 3.73NGIDD314 pKa = 3.25FGYY317 pKa = 11.01YY318 pKa = 8.69VFHH321 pKa = 7.66YY322 pKa = 10.39EE323 pKa = 4.68DD324 pKa = 3.95GRR326 pKa = 11.84NVIAPIEE333 pKa = 3.91LAGSIISGTEE343 pKa = 3.29LRR345 pKa = 11.84VRR347 pKa = 11.84LDD349 pKa = 3.19LATYY353 pKa = 6.96WTEE356 pKa = 4.13SPDD359 pKa = 3.24PRR361 pKa = 11.84LSYY364 pKa = 10.93FEE366 pKa = 4.47INLYY370 pKa = 10.15QGTLISEE377 pKa = 4.53DD378 pKa = 3.04GVYY381 pKa = 10.6FDD383 pKa = 4.44GHH385 pKa = 6.54APSSQIKK392 pKa = 9.75YY393 pKa = 9.9FNIWTT398 pKa = 3.82
Molecular weight: 44.56 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A7D7F5V9|A0A7D7F5V9_9CAUD Structural protein OS=Flavobacterium phage vB_FspP_elemoA_3-5A OX=2743819 GN=elemo35A_phanotate54 PE=4 SV=1
MM1 pKa = 6.93QRR3 pKa = 11.84CLSLSVFGSIRR14 pKa = 11.84NKK16 pKa = 10.73LNGSSFSLKK25 pKa = 9.63IVRR28 pKa = 11.84IAQLVRR34 pKa = 11.84AIDD37 pKa = 3.48SS38 pKa = 3.57
MM1 pKa = 6.93QRR3 pKa = 11.84CLSLSVFGSIRR14 pKa = 11.84NKK16 pKa = 10.73LNGSSFSLKK25 pKa = 9.63IVRR28 pKa = 11.84IAQLVRR34 pKa = 11.84AIDD37 pKa = 3.48SS38 pKa = 3.57
Molecular weight: 4.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
19272 |
30 |
3694 |
214.1 |
24.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.36 ± 0.406 | 0.711 ± 0.153 |
6.844 ± 0.223 | 7.363 ± 0.34 |
4.545 ± 0.238 | 5.936 ± 0.239 |
1.147 ± 0.156 | 7.752 ± 0.28 |
9.293 ± 0.671 | 7.887 ± 0.238 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.273 ± 0.176 | 7.207 ± 0.196 |
2.703 ± 0.125 | 3.056 ± 0.27 |
3.492 ± 0.182 | 7.244 ± 0.263 |
5.682 ± 0.308 | 5.998 ± 0.226 |
0.887 ± 0.111 | 4.618 ± 0.32 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
