
Mojiang virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Orthoparamyxovirinae; Henipavirus; Mojiang henipavirus
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|W8TIR0|W8TIR0_9MONO Isoform of W8SKS6 V protein OS=Mojiang virus OX=1474807 GN=C PE=4 SV=1
MM1 pKa = 7.94SYY3 pKa = 10.83EE4 pKa = 4.27DD5 pKa = 5.22RR6 pKa = 11.84IKK8 pKa = 10.62QIQNGLQIVDD18 pKa = 3.8LVKK21 pKa = 10.46KK22 pKa = 10.17VRR24 pKa = 11.84QEE26 pKa = 4.24SIEE29 pKa = 4.0KK30 pKa = 8.15PTYY33 pKa = 8.89GRR35 pKa = 11.84SAIGLPTTKK44 pKa = 10.36DD45 pKa = 2.84RR46 pKa = 11.84AAAWEE51 pKa = 4.32LFHH54 pKa = 7.56KK55 pKa = 10.8SSLDD59 pKa = 3.39EE60 pKa = 4.88AGPEE64 pKa = 3.85EE65 pKa = 4.91LPLKK69 pKa = 10.66EE70 pKa = 4.89GDD72 pKa = 4.36DD73 pKa = 4.04PADD76 pKa = 3.45TRR78 pKa = 11.84DD79 pKa = 3.49GVGIPEE85 pKa = 4.5PLHH88 pKa = 6.47NVDD91 pKa = 3.64SGGSRR96 pKa = 11.84TYY98 pKa = 11.05KK99 pKa = 8.96EE100 pKa = 4.56ANWDD104 pKa = 3.72EE105 pKa = 4.07GDD107 pKa = 3.85EE108 pKa = 4.87PILEE112 pKa = 4.33NQLVTNIQPNDD123 pKa = 3.92PGRR126 pKa = 11.84KK127 pKa = 5.62TAYY130 pKa = 9.7GKK132 pKa = 10.28SDD134 pKa = 3.17HH135 pKa = 6.92TYY137 pKa = 11.49ADD139 pKa = 3.45STNNRR144 pKa = 11.84TKK146 pKa = 10.75GGEE149 pKa = 3.8WSNGCSSPNKK159 pKa = 9.55VDD161 pKa = 3.16VSGIFQGDD169 pKa = 3.67VPKK172 pKa = 10.37DD173 pKa = 3.31TDD175 pKa = 3.75AEE177 pKa = 4.74GPSEE181 pKa = 4.12KK182 pKa = 9.84PKK184 pKa = 10.97SSFRR188 pKa = 11.84MNPNAQEE195 pKa = 4.09YY196 pKa = 9.81IPRR199 pKa = 11.84DD200 pKa = 3.77LTPLTVIIDD209 pKa = 3.84SCPFDD214 pKa = 5.0HH215 pKa = 6.99NHH217 pKa = 6.99ADD219 pKa = 5.2DD220 pKa = 5.03DD221 pKa = 5.42SEE223 pKa = 4.91DD224 pKa = 3.61QTEE227 pKa = 3.97DD228 pKa = 2.94SAYY231 pKa = 10.75VFEE234 pKa = 6.11AGLNKK239 pKa = 9.88PAVKK243 pKa = 9.96PRR245 pKa = 11.84MIKK248 pKa = 10.15AAQKK252 pKa = 7.82EE253 pKa = 4.47TLVDD257 pKa = 3.39QDD259 pKa = 4.2GEE261 pKa = 4.54VMNLSILPKK270 pKa = 9.41QRR272 pKa = 11.84KK273 pKa = 9.37SILNKK278 pKa = 10.28PIGAEE283 pKa = 3.65DD284 pKa = 5.6AIPKK288 pKa = 9.16KK289 pKa = 8.14QARR292 pKa = 11.84PSLVVIEE299 pKa = 5.24EE300 pKa = 4.17DD301 pKa = 5.71DD302 pKa = 3.96EE303 pKa = 4.45DD304 pKa = 4.07QKK306 pKa = 11.69SEE308 pKa = 4.43PIEE311 pKa = 4.86NIDD314 pKa = 4.48KK315 pKa = 10.96SDD317 pKa = 3.67AGSDD321 pKa = 3.02ITIFDD326 pKa = 3.99IADD329 pKa = 3.57KK330 pKa = 10.46ATDD333 pKa = 3.43HH334 pKa = 6.85LRR336 pKa = 11.84RR337 pKa = 11.84NQMAVKK343 pKa = 10.32AIGNIIEE350 pKa = 4.29TSAGVPIIQEE360 pKa = 3.97EE361 pKa = 4.6VIYY364 pKa = 10.88LSDD367 pKa = 4.97RR368 pKa = 11.84PTQPAPEE375 pKa = 5.01KK376 pKa = 10.53IPAKK380 pKa = 9.94EE381 pKa = 3.98SRR383 pKa = 11.84KK384 pKa = 9.81LRR386 pKa = 11.84ALEE389 pKa = 3.87KK390 pKa = 9.2TEE392 pKa = 4.05SGKK395 pKa = 11.21GEE397 pKa = 4.13SDD399 pKa = 3.02SGLRR403 pKa = 11.84LVKK406 pKa = 10.14KK407 pKa = 8.05GHH409 pKa = 5.47RR410 pKa = 11.84RR411 pKa = 11.84EE412 pKa = 5.01YY413 pKa = 10.01SLCWDD418 pKa = 3.3GSEE421 pKa = 3.92IKK423 pKa = 10.65VEE425 pKa = 4.33EE426 pKa = 4.33WCNPICSKK434 pKa = 10.85VKK436 pKa = 10.51SEE438 pKa = 4.12PSRR441 pKa = 11.84EE442 pKa = 3.86KK443 pKa = 10.09CTCKK447 pKa = 9.98QCPIMCQDD455 pKa = 3.29EE456 pKa = 4.68HH457 pKa = 5.98CTKK460 pKa = 10.17IEE462 pKa = 4.01YY463 pKa = 8.82EE464 pKa = 4.05
MM1 pKa = 7.94SYY3 pKa = 10.83EE4 pKa = 4.27DD5 pKa = 5.22RR6 pKa = 11.84IKK8 pKa = 10.62QIQNGLQIVDD18 pKa = 3.8LVKK21 pKa = 10.46KK22 pKa = 10.17VRR24 pKa = 11.84QEE26 pKa = 4.24SIEE29 pKa = 4.0KK30 pKa = 8.15PTYY33 pKa = 8.89GRR35 pKa = 11.84SAIGLPTTKK44 pKa = 10.36DD45 pKa = 2.84RR46 pKa = 11.84AAAWEE51 pKa = 4.32LFHH54 pKa = 7.56KK55 pKa = 10.8SSLDD59 pKa = 3.39EE60 pKa = 4.88AGPEE64 pKa = 3.85EE65 pKa = 4.91LPLKK69 pKa = 10.66EE70 pKa = 4.89GDD72 pKa = 4.36DD73 pKa = 4.04PADD76 pKa = 3.45TRR78 pKa = 11.84DD79 pKa = 3.49GVGIPEE85 pKa = 4.5PLHH88 pKa = 6.47NVDD91 pKa = 3.64SGGSRR96 pKa = 11.84TYY98 pKa = 11.05KK99 pKa = 8.96EE100 pKa = 4.56ANWDD104 pKa = 3.72EE105 pKa = 4.07GDD107 pKa = 3.85EE108 pKa = 4.87PILEE112 pKa = 4.33NQLVTNIQPNDD123 pKa = 3.92PGRR126 pKa = 11.84KK127 pKa = 5.62TAYY130 pKa = 9.7GKK132 pKa = 10.28SDD134 pKa = 3.17HH135 pKa = 6.92TYY137 pKa = 11.49ADD139 pKa = 3.45STNNRR144 pKa = 11.84TKK146 pKa = 10.75GGEE149 pKa = 3.8WSNGCSSPNKK159 pKa = 9.55VDD161 pKa = 3.16VSGIFQGDD169 pKa = 3.67VPKK172 pKa = 10.37DD173 pKa = 3.31TDD175 pKa = 3.75AEE177 pKa = 4.74GPSEE181 pKa = 4.12KK182 pKa = 9.84PKK184 pKa = 10.97SSFRR188 pKa = 11.84MNPNAQEE195 pKa = 4.09YY196 pKa = 9.81IPRR199 pKa = 11.84DD200 pKa = 3.77LTPLTVIIDD209 pKa = 3.84SCPFDD214 pKa = 5.0HH215 pKa = 6.99NHH217 pKa = 6.99ADD219 pKa = 5.2DD220 pKa = 5.03DD221 pKa = 5.42SEE223 pKa = 4.91DD224 pKa = 3.61QTEE227 pKa = 3.97DD228 pKa = 2.94SAYY231 pKa = 10.75VFEE234 pKa = 6.11AGLNKK239 pKa = 9.88PAVKK243 pKa = 9.96PRR245 pKa = 11.84MIKK248 pKa = 10.15AAQKK252 pKa = 7.82EE253 pKa = 4.47TLVDD257 pKa = 3.39QDD259 pKa = 4.2GEE261 pKa = 4.54VMNLSILPKK270 pKa = 9.41QRR272 pKa = 11.84KK273 pKa = 9.37SILNKK278 pKa = 10.28PIGAEE283 pKa = 3.65DD284 pKa = 5.6AIPKK288 pKa = 9.16KK289 pKa = 8.14QARR292 pKa = 11.84PSLVVIEE299 pKa = 5.24EE300 pKa = 4.17DD301 pKa = 5.71DD302 pKa = 3.96EE303 pKa = 4.45DD304 pKa = 4.07QKK306 pKa = 11.69SEE308 pKa = 4.43PIEE311 pKa = 4.86NIDD314 pKa = 4.48KK315 pKa = 10.96SDD317 pKa = 3.67AGSDD321 pKa = 3.02ITIFDD326 pKa = 3.99IADD329 pKa = 3.57KK330 pKa = 10.46ATDD333 pKa = 3.43HH334 pKa = 6.85LRR336 pKa = 11.84RR337 pKa = 11.84NQMAVKK343 pKa = 10.32AIGNIIEE350 pKa = 4.29TSAGVPIIQEE360 pKa = 3.97EE361 pKa = 4.6VIYY364 pKa = 10.88LSDD367 pKa = 4.97RR368 pKa = 11.84PTQPAPEE375 pKa = 5.01KK376 pKa = 10.53IPAKK380 pKa = 9.94EE381 pKa = 3.98SRR383 pKa = 11.84KK384 pKa = 9.81LRR386 pKa = 11.84ALEE389 pKa = 3.87KK390 pKa = 9.2TEE392 pKa = 4.05SGKK395 pKa = 11.21GEE397 pKa = 4.13SDD399 pKa = 3.02SGLRR403 pKa = 11.84LVKK406 pKa = 10.14KK407 pKa = 8.05GHH409 pKa = 5.47RR410 pKa = 11.84RR411 pKa = 11.84EE412 pKa = 5.01YY413 pKa = 10.01SLCWDD418 pKa = 3.3GSEE421 pKa = 3.92IKK423 pKa = 10.65VEE425 pKa = 4.33EE426 pKa = 4.33WCNPICSKK434 pKa = 10.85VKK436 pKa = 10.51SEE438 pKa = 4.12PSRR441 pKa = 11.84EE442 pKa = 3.86KK443 pKa = 10.09CTCKK447 pKa = 9.98QCPIMCQDD455 pKa = 3.29EE456 pKa = 4.68HH457 pKa = 5.98CTKK460 pKa = 10.17IEE462 pKa = 4.01YY463 pKa = 8.82EE464 pKa = 4.05
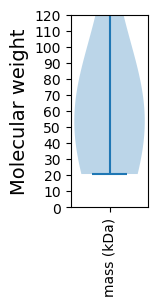
Molecular weight: 51.57 kDa
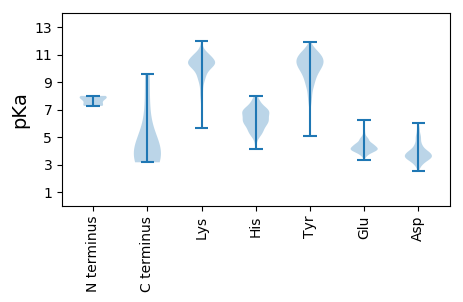
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W8SYF7|W8SYF7_9MONO Isoform of W8SKS6 W protein OS=Mojiang virus OX=1474807 GN=C PE=4 SV=1
MM1 pKa = 7.99ASRR4 pKa = 11.84LLTWSRR10 pKa = 11.84KK11 pKa = 8.83LGRR14 pKa = 11.84NRR16 pKa = 11.84SKK18 pKa = 10.57SQHH21 pKa = 5.06TGEE24 pKa = 4.19VQLAFQQPKK33 pKa = 9.84IEE35 pKa = 4.03QQPGSCSINLHH46 pKa = 5.79LMKK49 pKa = 10.05PDD51 pKa = 4.7LKK53 pKa = 11.14NCLLKK58 pKa = 10.74KK59 pKa = 10.66EE60 pKa = 4.2MTQLTLEE67 pKa = 4.41MEE69 pKa = 4.32WAFQSLFIMLTVEE82 pKa = 4.17EE83 pKa = 4.68AEE85 pKa = 4.1HH86 pKa = 6.09TKK88 pKa = 10.52RR89 pKa = 11.84QTGMRR94 pKa = 11.84EE95 pKa = 3.66TNQYY99 pKa = 11.34SKK101 pKa = 10.77ISWLQTFNRR110 pKa = 11.84MIQEE114 pKa = 3.89GRR116 pKa = 11.84LPTGNLITRR125 pKa = 11.84MLTAQIIEE133 pKa = 4.23PKK135 pKa = 9.23EE136 pKa = 3.72VSGAMDD142 pKa = 3.51VLRR145 pKa = 11.84LIRR148 pKa = 11.84LMCPAYY154 pKa = 10.5SRR156 pKa = 11.84AMYY159 pKa = 10.55LRR161 pKa = 11.84TQMLKK166 pKa = 10.86DD167 pKa = 3.54LVRR170 pKa = 11.84NPSHH174 pKa = 6.81HH175 pKa = 6.86SEE177 pKa = 3.97
MM1 pKa = 7.99ASRR4 pKa = 11.84LLTWSRR10 pKa = 11.84KK11 pKa = 8.83LGRR14 pKa = 11.84NRR16 pKa = 11.84SKK18 pKa = 10.57SQHH21 pKa = 5.06TGEE24 pKa = 4.19VQLAFQQPKK33 pKa = 9.84IEE35 pKa = 4.03QQPGSCSINLHH46 pKa = 5.79LMKK49 pKa = 10.05PDD51 pKa = 4.7LKK53 pKa = 11.14NCLLKK58 pKa = 10.74KK59 pKa = 10.66EE60 pKa = 4.2MTQLTLEE67 pKa = 4.41MEE69 pKa = 4.32WAFQSLFIMLTVEE82 pKa = 4.17EE83 pKa = 4.68AEE85 pKa = 4.1HH86 pKa = 6.09TKK88 pKa = 10.52RR89 pKa = 11.84QTGMRR94 pKa = 11.84EE95 pKa = 3.66TNQYY99 pKa = 11.34SKK101 pKa = 10.77ISWLQTFNRR110 pKa = 11.84MIQEE114 pKa = 3.89GRR116 pKa = 11.84LPTGNLITRR125 pKa = 11.84MLTAQIIEE133 pKa = 4.23PKK135 pKa = 9.23EE136 pKa = 3.72VSGAMDD142 pKa = 3.51VLRR145 pKa = 11.84LIRR148 pKa = 11.84LMCPAYY154 pKa = 10.5SRR156 pKa = 11.84AMYY159 pKa = 10.55LRR161 pKa = 11.84TQMLKK166 pKa = 10.86DD167 pKa = 3.54LVRR170 pKa = 11.84NPSHH174 pKa = 6.81HH175 pKa = 6.86SEE177 pKa = 3.97
Molecular weight: 20.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
6095 |
177 |
2277 |
677.2 |
76.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.595 ± 0.708 | 1.575 ± 0.318 |
6.497 ± 0.53 | 6.202 ± 0.634 |
2.97 ± 0.426 | 5.628 ± 0.46 |
1.854 ± 0.337 | 8.236 ± 0.47 |
6.776 ± 0.45 | 9.04 ± 0.734 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.592 ± 0.331 | 5.742 ± 0.271 |
4.742 ± 0.44 | 3.741 ± 0.384 |
4.791 ± 0.369 | 8.023 ± 0.273 |
5.89 ± 0.381 | 5.513 ± 0.375 |
1.017 ± 0.155 | 3.577 ± 0.509 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
