
Xinzhou nematode virus 7
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 9.06
Get precalculated fractions of proteins
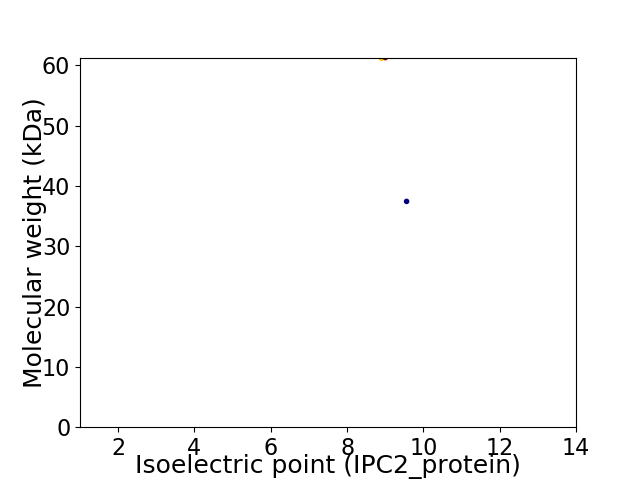
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KH95|A0A1L3KH95_9VIRU RNA-directed RNA polymerase OS=Xinzhou nematode virus 7 OX=1923775 PE=4 SV=1
MM1 pKa = 7.45GLGASAMVAMHH12 pKa = 6.61SSVDD16 pKa = 3.75MQTSMPTAGVVIGGVISRR34 pKa = 11.84GPFKK38 pKa = 10.39PLRR41 pKa = 11.84VGATIRR47 pKa = 11.84YY48 pKa = 8.08GYY50 pKa = 9.58YY51 pKa = 10.2GRR53 pKa = 11.84RR54 pKa = 11.84PQSTRR59 pKa = 11.84IQSLFPDD66 pKa = 3.95VEE68 pKa = 4.23GLLPLEE74 pKa = 4.16TRR76 pKa = 11.84TVNVHH81 pKa = 5.32LVCWEE86 pKa = 3.93NEE88 pKa = 4.14IVCLRR93 pKa = 11.84NRR95 pKa = 11.84VLMQTPQPRR104 pKa = 11.84PHH106 pKa = 7.77SMGRR110 pKa = 11.84LQLHH114 pKa = 5.58GHH116 pKa = 6.4ALCSIVLSFIGRR128 pKa = 11.84LTPWSLSKK136 pKa = 9.52TLRR139 pKa = 11.84HH140 pKa = 4.76MVVRR144 pKa = 11.84HH145 pKa = 5.42GAALYY150 pKa = 10.39VPAAEE155 pKa = 4.76SLRR158 pKa = 11.84QEE160 pKa = 4.11PLGSIDD166 pKa = 3.6ARR168 pKa = 11.84VSMFLKK174 pKa = 10.05VEE176 pKa = 4.14KK177 pKa = 10.9VFAEE181 pKa = 4.55DD182 pKa = 3.57PAEE185 pKa = 4.23KK186 pKa = 10.23APRR189 pKa = 11.84AIQYY193 pKa = 8.77RR194 pKa = 11.84GPRR197 pKa = 11.84FNLVLGRR204 pKa = 11.84FILAYY209 pKa = 10.15EE210 pKa = 3.96EE211 pKa = 3.83AFYY214 pKa = 10.89RR215 pKa = 11.84AFNHH219 pKa = 5.42VNASRR224 pKa = 11.84CHH226 pKa = 5.47TSKK229 pKa = 11.16GLTPDD234 pKa = 3.17ARR236 pKa = 11.84AALLWEE242 pKa = 4.68LWSKK246 pKa = 10.68NPRR249 pKa = 11.84PAALNVDD256 pKa = 3.91ASRR259 pKa = 11.84FDD261 pKa = 3.23AHH263 pKa = 5.69VTVEE267 pKa = 4.0MLLLEE272 pKa = 4.14SLYY275 pKa = 11.03YY276 pKa = 10.41EE277 pKa = 4.43SSFPNCRR284 pKa = 11.84LLRR287 pKa = 11.84WLLRR291 pKa = 11.84CQRR294 pKa = 11.84RR295 pKa = 11.84NRR297 pKa = 11.84GCGKK301 pKa = 10.52FGTTYY306 pKa = 8.58TLKK309 pKa = 10.15GGRR312 pKa = 11.84MSGDD316 pKa = 3.13VNTALGNTLIQMTVLSFLAGRR337 pKa = 11.84SFDD340 pKa = 4.12LVVEE344 pKa = 4.49GDD346 pKa = 3.68DD347 pKa = 3.82AVIFGDD353 pKa = 3.34VDD355 pKa = 4.59DD356 pKa = 4.17ITRR359 pKa = 11.84LEE361 pKa = 3.89AVITSKK367 pKa = 11.23ALAVGFQLKK376 pKa = 9.88VSKK379 pKa = 10.6ALYY382 pKa = 10.24LEE384 pKa = 3.86QLEE387 pKa = 4.58YY388 pKa = 11.09CSTRR392 pKa = 11.84VIRR395 pKa = 11.84SQSGDD400 pKa = 2.95VRR402 pKa = 11.84AIRR405 pKa = 11.84EE406 pKa = 4.32WPRR409 pKa = 11.84PLAMDD414 pKa = 4.66RR415 pKa = 11.84YY416 pKa = 7.27TVKK419 pKa = 10.28IVSGQRR425 pKa = 11.84ATNRR429 pKa = 11.84KK430 pKa = 7.88ARR432 pKa = 11.84TMAVCFMSLYY442 pKa = 10.55SGLPVYY448 pKa = 9.63EE449 pKa = 3.88EE450 pKa = 3.76WARR453 pKa = 11.84YY454 pKa = 8.24LLSWSDD460 pKa = 3.2PSFGVDD466 pKa = 3.06EE467 pKa = 4.57SQDD470 pKa = 3.09RR471 pKa = 11.84RR472 pKa = 11.84LWFIAAEE479 pKa = 4.23ACVGTGDD486 pKa = 4.31QVPADD491 pKa = 3.89GLRR494 pKa = 11.84SSFYY498 pKa = 9.36MATTIEE504 pKa = 4.28PSEE507 pKa = 3.99QLRR510 pKa = 11.84VEE512 pKa = 4.11GLLRR516 pKa = 11.84DD517 pKa = 3.51QRR519 pKa = 11.84GPHH522 pKa = 6.47PAYY525 pKa = 9.82ILGGEE530 pKa = 4.04LRR532 pKa = 11.84ALRR535 pKa = 11.84RR536 pKa = 11.84AAAQLQNSAPSSS548 pKa = 3.56
MM1 pKa = 7.45GLGASAMVAMHH12 pKa = 6.61SSVDD16 pKa = 3.75MQTSMPTAGVVIGGVISRR34 pKa = 11.84GPFKK38 pKa = 10.39PLRR41 pKa = 11.84VGATIRR47 pKa = 11.84YY48 pKa = 8.08GYY50 pKa = 9.58YY51 pKa = 10.2GRR53 pKa = 11.84RR54 pKa = 11.84PQSTRR59 pKa = 11.84IQSLFPDD66 pKa = 3.95VEE68 pKa = 4.23GLLPLEE74 pKa = 4.16TRR76 pKa = 11.84TVNVHH81 pKa = 5.32LVCWEE86 pKa = 3.93NEE88 pKa = 4.14IVCLRR93 pKa = 11.84NRR95 pKa = 11.84VLMQTPQPRR104 pKa = 11.84PHH106 pKa = 7.77SMGRR110 pKa = 11.84LQLHH114 pKa = 5.58GHH116 pKa = 6.4ALCSIVLSFIGRR128 pKa = 11.84LTPWSLSKK136 pKa = 9.52TLRR139 pKa = 11.84HH140 pKa = 4.76MVVRR144 pKa = 11.84HH145 pKa = 5.42GAALYY150 pKa = 10.39VPAAEE155 pKa = 4.76SLRR158 pKa = 11.84QEE160 pKa = 4.11PLGSIDD166 pKa = 3.6ARR168 pKa = 11.84VSMFLKK174 pKa = 10.05VEE176 pKa = 4.14KK177 pKa = 10.9VFAEE181 pKa = 4.55DD182 pKa = 3.57PAEE185 pKa = 4.23KK186 pKa = 10.23APRR189 pKa = 11.84AIQYY193 pKa = 8.77RR194 pKa = 11.84GPRR197 pKa = 11.84FNLVLGRR204 pKa = 11.84FILAYY209 pKa = 10.15EE210 pKa = 3.96EE211 pKa = 3.83AFYY214 pKa = 10.89RR215 pKa = 11.84AFNHH219 pKa = 5.42VNASRR224 pKa = 11.84CHH226 pKa = 5.47TSKK229 pKa = 11.16GLTPDD234 pKa = 3.17ARR236 pKa = 11.84AALLWEE242 pKa = 4.68LWSKK246 pKa = 10.68NPRR249 pKa = 11.84PAALNVDD256 pKa = 3.91ASRR259 pKa = 11.84FDD261 pKa = 3.23AHH263 pKa = 5.69VTVEE267 pKa = 4.0MLLLEE272 pKa = 4.14SLYY275 pKa = 11.03YY276 pKa = 10.41EE277 pKa = 4.43SSFPNCRR284 pKa = 11.84LLRR287 pKa = 11.84WLLRR291 pKa = 11.84CQRR294 pKa = 11.84RR295 pKa = 11.84NRR297 pKa = 11.84GCGKK301 pKa = 10.52FGTTYY306 pKa = 8.58TLKK309 pKa = 10.15GGRR312 pKa = 11.84MSGDD316 pKa = 3.13VNTALGNTLIQMTVLSFLAGRR337 pKa = 11.84SFDD340 pKa = 4.12LVVEE344 pKa = 4.49GDD346 pKa = 3.68DD347 pKa = 3.82AVIFGDD353 pKa = 3.34VDD355 pKa = 4.59DD356 pKa = 4.17ITRR359 pKa = 11.84LEE361 pKa = 3.89AVITSKK367 pKa = 11.23ALAVGFQLKK376 pKa = 9.88VSKK379 pKa = 10.6ALYY382 pKa = 10.24LEE384 pKa = 3.86QLEE387 pKa = 4.58YY388 pKa = 11.09CSTRR392 pKa = 11.84VIRR395 pKa = 11.84SQSGDD400 pKa = 2.95VRR402 pKa = 11.84AIRR405 pKa = 11.84EE406 pKa = 4.32WPRR409 pKa = 11.84PLAMDD414 pKa = 4.66RR415 pKa = 11.84YY416 pKa = 7.27TVKK419 pKa = 10.28IVSGQRR425 pKa = 11.84ATNRR429 pKa = 11.84KK430 pKa = 7.88ARR432 pKa = 11.84TMAVCFMSLYY442 pKa = 10.55SGLPVYY448 pKa = 9.63EE449 pKa = 3.88EE450 pKa = 3.76WARR453 pKa = 11.84YY454 pKa = 8.24LLSWSDD460 pKa = 3.2PSFGVDD466 pKa = 3.06EE467 pKa = 4.57SQDD470 pKa = 3.09RR471 pKa = 11.84RR472 pKa = 11.84LWFIAAEE479 pKa = 4.23ACVGTGDD486 pKa = 4.31QVPADD491 pKa = 3.89GLRR494 pKa = 11.84SSFYY498 pKa = 9.36MATTIEE504 pKa = 4.28PSEE507 pKa = 3.99QLRR510 pKa = 11.84VEE512 pKa = 4.11GLLRR516 pKa = 11.84DD517 pKa = 3.51QRR519 pKa = 11.84GPHH522 pKa = 6.47PAYY525 pKa = 9.82ILGGEE530 pKa = 4.04LRR532 pKa = 11.84ALRR535 pKa = 11.84RR536 pKa = 11.84AAAQLQNSAPSSS548 pKa = 3.56
Molecular weight: 61.16 kDa
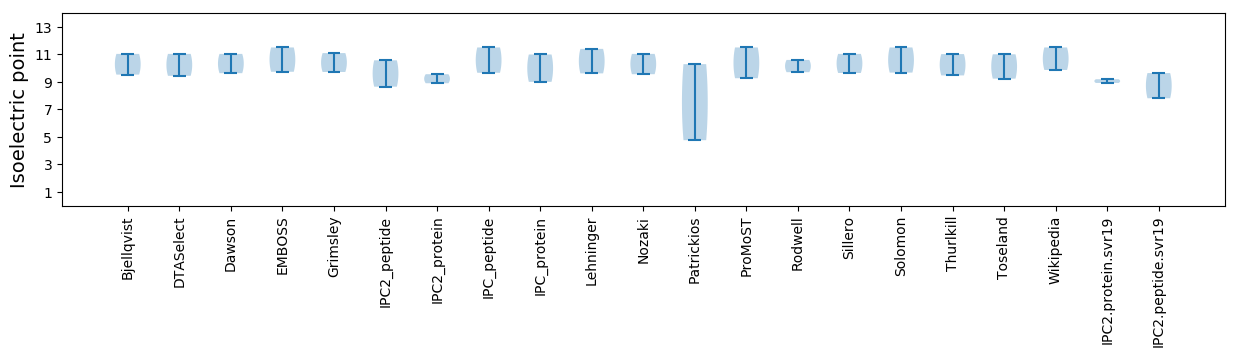
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KH95|A0A1L3KH95_9VIRU RNA-directed RNA polymerase OS=Xinzhou nematode virus 7 OX=1923775 PE=4 SV=1
MM1 pKa = 7.87DD2 pKa = 4.5NEE4 pKa = 4.19MLRR7 pKa = 11.84GGRR10 pKa = 11.84GPPINQPRR18 pKa = 11.84APPPDD23 pKa = 3.27RR24 pKa = 11.84RR25 pKa = 11.84GGAGRR30 pKa = 11.84RR31 pKa = 11.84TRR33 pKa = 11.84NQVGADD39 pKa = 3.15QLGFRR44 pKa = 11.84RR45 pKa = 11.84GPRR48 pKa = 11.84TQLRR52 pKa = 11.84GPSRR56 pKa = 11.84VRR58 pKa = 11.84SSRR61 pKa = 11.84ASVARR66 pKa = 11.84GGRR69 pKa = 11.84GNFPPPPPRR78 pKa = 11.84GPAQFEE84 pKa = 4.14ARR86 pKa = 11.84GMPHH90 pKa = 6.45HH91 pKa = 7.11RR92 pKa = 11.84GSQGLPRR99 pKa = 11.84PLPAPGQQQAAPVDD113 pKa = 4.03HH114 pKa = 7.07FPPLVPRR121 pKa = 11.84EE122 pKa = 4.39GEE124 pKa = 4.04PGPQPVLPCVVQWPCVPAPPGPAGPQAQPPAPPGAVNCGAPQRR167 pKa = 11.84HH168 pKa = 5.09RR169 pKa = 11.84VAPIRR174 pKa = 11.84ALSEE178 pKa = 3.91GGEE181 pKa = 4.08PFEE184 pKa = 4.88PEE186 pKa = 3.82VGPTLRR192 pKa = 11.84DD193 pKa = 2.89RR194 pKa = 11.84CRR196 pKa = 11.84EE197 pKa = 3.89YY198 pKa = 11.03LSRR201 pKa = 11.84RR202 pKa = 11.84LVWLARR208 pKa = 11.84TPEE211 pKa = 3.49NRR213 pKa = 11.84AAVVRR218 pKa = 11.84AARR221 pKa = 11.84TFVSSQDD228 pKa = 3.28PDD230 pKa = 4.05CLVRR234 pKa = 11.84APWLSALIDD243 pKa = 4.0EE244 pKa = 4.97LWFDD248 pKa = 4.22LRR250 pKa = 11.84VEE252 pKa = 4.24LEE254 pKa = 4.17LGRR257 pKa = 11.84VRR259 pKa = 11.84PWSVALHH266 pKa = 5.73EE267 pKa = 5.0ARR269 pKa = 11.84HH270 pKa = 4.5AHH272 pKa = 4.67YY273 pKa = 11.11VEE275 pKa = 4.69GRR277 pKa = 11.84AHH279 pKa = 6.89EE280 pKa = 5.35LFTPWRR286 pKa = 11.84LLNYY290 pKa = 7.44EE291 pKa = 3.98QSLGLDD297 pKa = 3.08QLSAYY302 pKa = 7.13WRR304 pKa = 11.84PIALCAGGAGLFCGAVAIGCWLGGAHH330 pKa = 6.68RR331 pKa = 11.84RR332 pKa = 11.84LPFEE336 pKa = 4.03VVLRR340 pKa = 11.84WGG342 pKa = 3.34
MM1 pKa = 7.87DD2 pKa = 4.5NEE4 pKa = 4.19MLRR7 pKa = 11.84GGRR10 pKa = 11.84GPPINQPRR18 pKa = 11.84APPPDD23 pKa = 3.27RR24 pKa = 11.84RR25 pKa = 11.84GGAGRR30 pKa = 11.84RR31 pKa = 11.84TRR33 pKa = 11.84NQVGADD39 pKa = 3.15QLGFRR44 pKa = 11.84RR45 pKa = 11.84GPRR48 pKa = 11.84TQLRR52 pKa = 11.84GPSRR56 pKa = 11.84VRR58 pKa = 11.84SSRR61 pKa = 11.84ASVARR66 pKa = 11.84GGRR69 pKa = 11.84GNFPPPPPRR78 pKa = 11.84GPAQFEE84 pKa = 4.14ARR86 pKa = 11.84GMPHH90 pKa = 6.45HH91 pKa = 7.11RR92 pKa = 11.84GSQGLPRR99 pKa = 11.84PLPAPGQQQAAPVDD113 pKa = 4.03HH114 pKa = 7.07FPPLVPRR121 pKa = 11.84EE122 pKa = 4.39GEE124 pKa = 4.04PGPQPVLPCVVQWPCVPAPPGPAGPQAQPPAPPGAVNCGAPQRR167 pKa = 11.84HH168 pKa = 5.09RR169 pKa = 11.84VAPIRR174 pKa = 11.84ALSEE178 pKa = 3.91GGEE181 pKa = 4.08PFEE184 pKa = 4.88PEE186 pKa = 3.82VGPTLRR192 pKa = 11.84DD193 pKa = 2.89RR194 pKa = 11.84CRR196 pKa = 11.84EE197 pKa = 3.89YY198 pKa = 11.03LSRR201 pKa = 11.84RR202 pKa = 11.84LVWLARR208 pKa = 11.84TPEE211 pKa = 3.49NRR213 pKa = 11.84AAVVRR218 pKa = 11.84AARR221 pKa = 11.84TFVSSQDD228 pKa = 3.28PDD230 pKa = 4.05CLVRR234 pKa = 11.84APWLSALIDD243 pKa = 4.0EE244 pKa = 4.97LWFDD248 pKa = 4.22LRR250 pKa = 11.84VEE252 pKa = 4.24LEE254 pKa = 4.17LGRR257 pKa = 11.84VRR259 pKa = 11.84PWSVALHH266 pKa = 5.73EE267 pKa = 5.0ARR269 pKa = 11.84HH270 pKa = 4.5AHH272 pKa = 4.67YY273 pKa = 11.11VEE275 pKa = 4.69GRR277 pKa = 11.84AHH279 pKa = 6.89EE280 pKa = 5.35LFTPWRR286 pKa = 11.84LLNYY290 pKa = 7.44EE291 pKa = 3.98QSLGLDD297 pKa = 3.08QLSAYY302 pKa = 7.13WRR304 pKa = 11.84PIALCAGGAGLFCGAVAIGCWLGGAHH330 pKa = 6.68RR331 pKa = 11.84RR332 pKa = 11.84LPFEE336 pKa = 4.03VVLRR340 pKa = 11.84WGG342 pKa = 3.34
Molecular weight: 37.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
890 |
342 |
548 |
445.0 |
49.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.663 ± 0.538 | 2.022 ± 0.197 |
3.596 ± 0.418 | 5.169 ± 0.059 |
3.371 ± 0.278 | 8.764 ± 1.462 |
2.247 ± 0.239 | 2.809 ± 0.839 |
1.573 ± 0.98 | 10.449 ± 0.863 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.135 ± 0.783 | 2.36 ± 0.195 |
8.652 ± 3.535 | 4.157 ± 0.507 |
11.011 ± 1.337 | 6.292 ± 1.551 |
3.708 ± 1.217 | 7.528 ± 0.318 |
2.022 ± 0.379 | 2.472 ± 0.811 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
