
Cyanophage S-RIM4
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; unclassified Myoviridae
Average proteome isoelectric point is 5.87
Get precalculated fractions of proteins
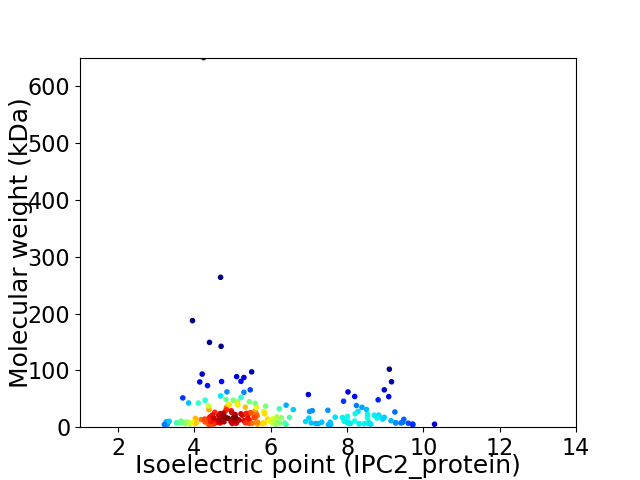
Virtual 2D-PAGE plot for 228 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
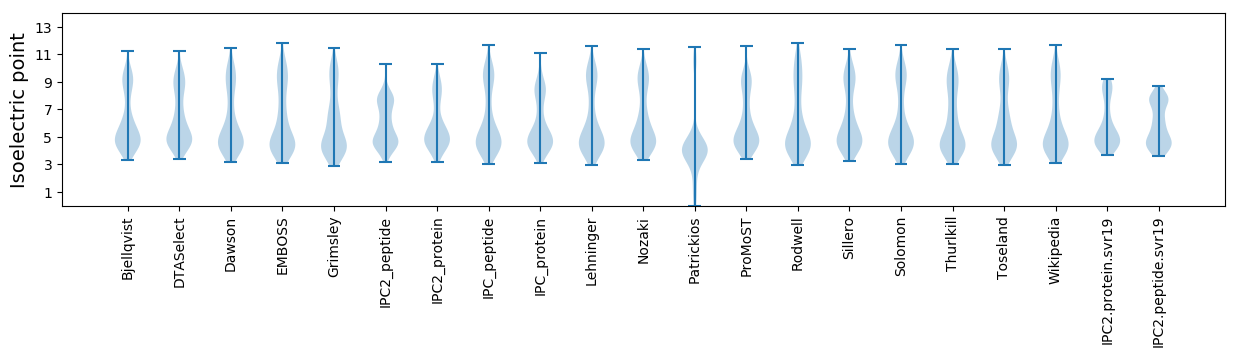
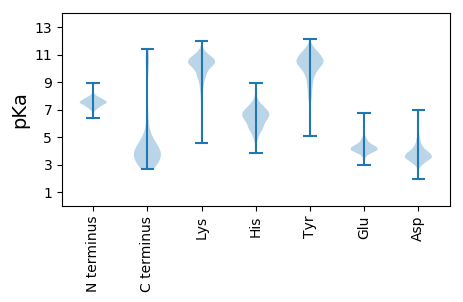
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A482MNX7|A0A482MNX7_9CAUD Tail fiber protein OS=Cyanophage S-RIM4 OX=2530169 GN=RW110999_045 PE=4 SV=1
MM1 pKa = 7.99ADD3 pKa = 3.41QGILAQSKK11 pKa = 8.97PAAATDD17 pKa = 3.72TVLYY21 pKa = 9.28RR22 pKa = 11.84APVDD26 pKa = 3.68QSASTVLNIANDD38 pKa = 4.41GTGSAYY44 pKa = 10.46SVAVKK49 pKa = 10.38DD50 pKa = 3.48YY51 pKa = 10.75DD52 pKa = 4.17QRR54 pKa = 11.84LTLDD58 pKa = 3.06ASTYY62 pKa = 10.32LLHH65 pKa = 7.08PGDD68 pKa = 4.96IISSYY73 pKa = 11.61SLVFDD78 pKa = 4.22VNMTASSGFTPGQLITTDD96 pKa = 3.8DD97 pKa = 4.03GEE99 pKa = 4.08KK100 pKa = 10.38SFRR103 pKa = 11.84FEE105 pKa = 3.92SFRR108 pKa = 11.84VPEE111 pKa = 3.9FTEE114 pKa = 3.71IFVKK118 pKa = 10.35IEE120 pKa = 4.11SIRR123 pKa = 11.84AITVEE128 pKa = 4.39SVSGTFTVGSTVSTGVAPDD147 pKa = 3.49DD148 pKa = 3.85TTAVIYY154 pKa = 10.28GVLDD158 pKa = 3.48QSTSAILYY166 pKa = 7.83VGPSTINGSGVEE178 pKa = 4.12FADD181 pKa = 4.08GDD183 pKa = 4.15SLAITGASGTISAGGIAAAADD204 pKa = 3.52EE205 pKa = 4.97FVFSITTAGGTYY217 pKa = 10.59NSFLTEE223 pKa = 4.05TTSLFNDD230 pKa = 3.17RR231 pKa = 11.84TYY233 pKa = 11.3RR234 pKa = 11.84FDD236 pKa = 3.71VSDD239 pKa = 3.77SSMTGSDD246 pKa = 3.34FALSFTANGIYY257 pKa = 10.66GPDD260 pKa = 3.58GAIGGVSPDD269 pKa = 4.32DD270 pKa = 3.28GTEE273 pKa = 3.95YY274 pKa = 11.3VFGQTVNGTPGSAGAYY290 pKa = 7.96IQYY293 pKa = 10.43DD294 pKa = 4.09FNAGATTNDD303 pKa = 3.61PVTGVPYY310 pKa = 10.47FGGSILYY317 pKa = 10.03FYY319 pKa = 10.95DD320 pKa = 3.46NGDD323 pKa = 3.57AAFGGSDD330 pKa = 3.33RR331 pKa = 11.84QFLINSSTEE340 pKa = 3.68FNEE343 pKa = 3.7FFIYY347 pKa = 10.44DD348 pKa = 3.83LEE350 pKa = 5.96GSLVASSDD358 pKa = 3.43TFEE361 pKa = 4.44VQGVTYY367 pKa = 9.53TLQTINAGPYY377 pKa = 8.8GYY379 pKa = 10.29VRR381 pKa = 11.84RR382 pKa = 11.84YY383 pKa = 10.43SGTTLDD389 pKa = 3.99VIKK392 pKa = 10.99GPGSQDD398 pKa = 3.03FAGTDD403 pKa = 3.69TIFDD407 pKa = 3.76NPKK410 pKa = 10.75DD411 pKa = 3.69GDD413 pKa = 3.7ATRR416 pKa = 11.84SLMTVSSVDD425 pKa = 3.43VASAAVTAEE434 pKa = 3.83NYY436 pKa = 8.32LTVGATNSANNVDD449 pKa = 4.86KK450 pKa = 10.4ITSLVVGPGEE460 pKa = 4.07VVVVNSTTQNNVFSLVGFEE479 pKa = 4.6DD480 pKa = 3.68QATSFPTRR488 pKa = 11.84VFDD491 pKa = 4.94FSS493 pKa = 3.53
MM1 pKa = 7.99ADD3 pKa = 3.41QGILAQSKK11 pKa = 8.97PAAATDD17 pKa = 3.72TVLYY21 pKa = 9.28RR22 pKa = 11.84APVDD26 pKa = 3.68QSASTVLNIANDD38 pKa = 4.41GTGSAYY44 pKa = 10.46SVAVKK49 pKa = 10.38DD50 pKa = 3.48YY51 pKa = 10.75DD52 pKa = 4.17QRR54 pKa = 11.84LTLDD58 pKa = 3.06ASTYY62 pKa = 10.32LLHH65 pKa = 7.08PGDD68 pKa = 4.96IISSYY73 pKa = 11.61SLVFDD78 pKa = 4.22VNMTASSGFTPGQLITTDD96 pKa = 3.8DD97 pKa = 4.03GEE99 pKa = 4.08KK100 pKa = 10.38SFRR103 pKa = 11.84FEE105 pKa = 3.92SFRR108 pKa = 11.84VPEE111 pKa = 3.9FTEE114 pKa = 3.71IFVKK118 pKa = 10.35IEE120 pKa = 4.11SIRR123 pKa = 11.84AITVEE128 pKa = 4.39SVSGTFTVGSTVSTGVAPDD147 pKa = 3.49DD148 pKa = 3.85TTAVIYY154 pKa = 10.28GVLDD158 pKa = 3.48QSTSAILYY166 pKa = 7.83VGPSTINGSGVEE178 pKa = 4.12FADD181 pKa = 4.08GDD183 pKa = 4.15SLAITGASGTISAGGIAAAADD204 pKa = 3.52EE205 pKa = 4.97FVFSITTAGGTYY217 pKa = 10.59NSFLTEE223 pKa = 4.05TTSLFNDD230 pKa = 3.17RR231 pKa = 11.84TYY233 pKa = 11.3RR234 pKa = 11.84FDD236 pKa = 3.71VSDD239 pKa = 3.77SSMTGSDD246 pKa = 3.34FALSFTANGIYY257 pKa = 10.66GPDD260 pKa = 3.58GAIGGVSPDD269 pKa = 4.32DD270 pKa = 3.28GTEE273 pKa = 3.95YY274 pKa = 11.3VFGQTVNGTPGSAGAYY290 pKa = 7.96IQYY293 pKa = 10.43DD294 pKa = 4.09FNAGATTNDD303 pKa = 3.61PVTGVPYY310 pKa = 10.47FGGSILYY317 pKa = 10.03FYY319 pKa = 10.95DD320 pKa = 3.46NGDD323 pKa = 3.57AAFGGSDD330 pKa = 3.33RR331 pKa = 11.84QFLINSSTEE340 pKa = 3.68FNEE343 pKa = 3.7FFIYY347 pKa = 10.44DD348 pKa = 3.83LEE350 pKa = 5.96GSLVASSDD358 pKa = 3.43TFEE361 pKa = 4.44VQGVTYY367 pKa = 9.53TLQTINAGPYY377 pKa = 8.8GYY379 pKa = 10.29VRR381 pKa = 11.84RR382 pKa = 11.84YY383 pKa = 10.43SGTTLDD389 pKa = 3.99VIKK392 pKa = 10.99GPGSQDD398 pKa = 3.03FAGTDD403 pKa = 3.69TIFDD407 pKa = 3.76NPKK410 pKa = 10.75DD411 pKa = 3.69GDD413 pKa = 3.7ATRR416 pKa = 11.84SLMTVSSVDD425 pKa = 3.43VASAAVTAEE434 pKa = 3.83NYY436 pKa = 8.32LTVGATNSANNVDD449 pKa = 4.86KK450 pKa = 10.4ITSLVVGPGEE460 pKa = 4.07VVVVNSTTQNNVFSLVGFEE479 pKa = 4.6DD480 pKa = 3.68QATSFPTRR488 pKa = 11.84VFDD491 pKa = 4.94FSS493 pKa = 3.53
Molecular weight: 51.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A482MN93|A0A482MN93_9CAUD Baseplate wedge protein OS=Cyanophage S-RIM4 OX=2530169 GN=RW110999_098 PE=4 SV=1
MM1 pKa = 7.59KK2 pKa = 10.38ALTKK6 pKa = 10.24RR7 pKa = 11.84RR8 pKa = 11.84SANYY12 pKa = 8.56FANQVKK18 pKa = 10.15VLLLILIGVLIYY30 pKa = 10.12QSKK33 pKa = 10.14DD34 pKa = 2.93ARR36 pKa = 11.84FFISDD41 pKa = 3.81RR42 pKa = 11.84LNDD45 pKa = 3.9ASEE48 pKa = 4.55MIRR51 pKa = 11.84PAPQFEE57 pKa = 3.85IRR59 pKa = 11.84YY60 pKa = 9.03
MM1 pKa = 7.59KK2 pKa = 10.38ALTKK6 pKa = 10.24RR7 pKa = 11.84RR8 pKa = 11.84SANYY12 pKa = 8.56FANQVKK18 pKa = 10.15VLLLILIGVLIYY30 pKa = 10.12QSKK33 pKa = 10.14DD34 pKa = 2.93ARR36 pKa = 11.84FFISDD41 pKa = 3.81RR42 pKa = 11.84LNDD45 pKa = 3.9ASEE48 pKa = 4.55MIRR51 pKa = 11.84PAPQFEE57 pKa = 3.85IRR59 pKa = 11.84YY60 pKa = 9.03
Molecular weight: 7.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
55635 |
37 |
6026 |
244.0 |
27.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.107 ± 0.212 | 0.944 ± 0.102 |
6.766 ± 0.126 | 6.167 ± 0.255 |
4.404 ± 0.114 | 7.531 ± 0.315 |
1.447 ± 0.137 | 6.311 ± 0.193 |
5.852 ± 0.429 | 7.299 ± 0.133 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.148 ± 0.224 | 5.782 ± 0.203 |
4.015 ± 0.134 | 3.726 ± 0.098 |
4.166 ± 0.151 | 6.73 ± 0.198 |
7.4 ± 0.352 | 6.751 ± 0.202 |
1.165 ± 0.071 | 4.289 ± 0.145 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
