
Sus scrofa polyomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Alphapolyomavirus
Average proteome isoelectric point is 7.03
Get precalculated fractions of proteins
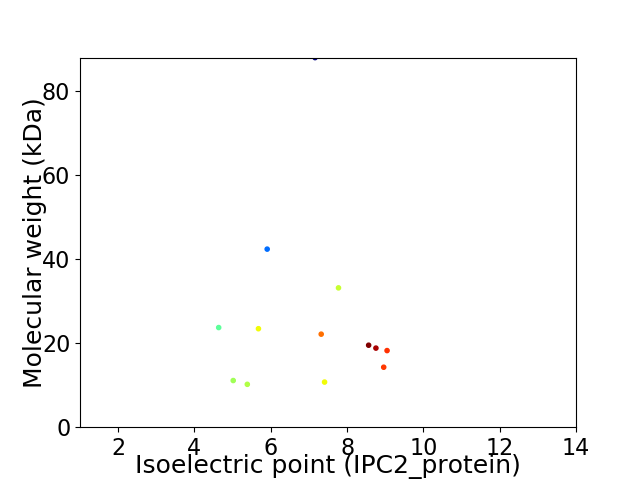
Virtual 2D-PAGE plot for 13 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A162GN61|A0A162GN61_9POLY Minor capsid protein VP2 OS=Sus scrofa polyomavirus 1 OX=1680894 GN=VP3 PE=3 SV=1
MM1 pKa = 7.62GGFLSLILDD10 pKa = 4.53LSTIAVDD17 pKa = 4.54LSASTGIALEE27 pKa = 4.83AILTGEE33 pKa = 4.08ALAALEE39 pKa = 4.48AEE41 pKa = 4.86VTALMTLEE49 pKa = 4.3GLSGVEE55 pKa = 4.72ALAQLGFTAEE65 pKa = 3.9QFANFSLVSSIVNQAVTYY83 pKa = 7.6GTIFQTVSGASALISAGIRR102 pKa = 11.84LGLEE106 pKa = 3.55EE107 pKa = 5.27RR108 pKa = 11.84SIVDD112 pKa = 4.59LNLHH116 pKa = 5.5GQGLTINGLSKK127 pKa = 10.09EE128 pKa = 4.89TITHH132 pKa = 5.65MASGFAFDD140 pKa = 4.15PFHH143 pKa = 6.42WSEE146 pKa = 4.4SLLHH150 pKa = 6.24SIGRR154 pKa = 11.84GINRR158 pKa = 11.84MTEE161 pKa = 3.89LPVNNNLARR170 pKa = 11.84LAEE173 pKa = 4.08EE174 pKa = 4.95GRR176 pKa = 11.84WKK178 pKa = 10.42LQRR181 pKa = 11.84EE182 pKa = 4.5GTDD185 pKa = 3.65DD186 pKa = 3.98GDD188 pKa = 3.93SGEE191 pKa = 4.53VIAMYY196 pKa = 8.7TPPGGSHH203 pKa = 6.39QRR205 pKa = 11.84VCPDD209 pKa = 2.66WLLPLVLGLSGSPRR223 pKa = 11.84GII225 pKa = 3.59
MM1 pKa = 7.62GGFLSLILDD10 pKa = 4.53LSTIAVDD17 pKa = 4.54LSASTGIALEE27 pKa = 4.83AILTGEE33 pKa = 4.08ALAALEE39 pKa = 4.48AEE41 pKa = 4.86VTALMTLEE49 pKa = 4.3GLSGVEE55 pKa = 4.72ALAQLGFTAEE65 pKa = 3.9QFANFSLVSSIVNQAVTYY83 pKa = 7.6GTIFQTVSGASALISAGIRR102 pKa = 11.84LGLEE106 pKa = 3.55EE107 pKa = 5.27RR108 pKa = 11.84SIVDD112 pKa = 4.59LNLHH116 pKa = 5.5GQGLTINGLSKK127 pKa = 10.09EE128 pKa = 4.89TITHH132 pKa = 5.65MASGFAFDD140 pKa = 4.15PFHH143 pKa = 6.42WSEE146 pKa = 4.4SLLHH150 pKa = 6.24SIGRR154 pKa = 11.84GINRR158 pKa = 11.84MTEE161 pKa = 3.89LPVNNNLARR170 pKa = 11.84LAEE173 pKa = 4.08EE174 pKa = 4.95GRR176 pKa = 11.84WKK178 pKa = 10.42LQRR181 pKa = 11.84EE182 pKa = 4.5GTDD185 pKa = 3.65DD186 pKa = 3.98GDD188 pKa = 3.93SGEE191 pKa = 4.53VIAMYY196 pKa = 8.7TPPGGSHH203 pKa = 6.39QRR205 pKa = 11.84VCPDD209 pKa = 2.66WLLPLVLGLSGSPRR223 pKa = 11.84GII225 pKa = 3.59
Molecular weight: 23.68 kDa
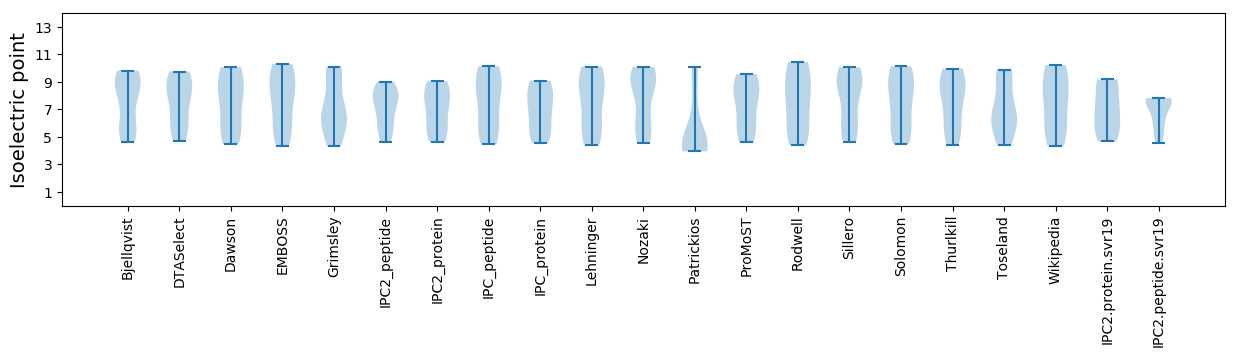
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A162GNC4|A0A162GNC4_9POLY 93 T antigen OS=Sus scrofa polyomavirus 1 OX=1680894 GN=93 TAg PE=4 SV=1
MM1 pKa = 8.08DD2 pKa = 4.21IALTRR7 pKa = 11.84EE8 pKa = 3.85EE9 pKa = 4.06RR10 pKa = 11.84KK11 pKa = 9.89EE12 pKa = 3.81LSEE15 pKa = 4.47LLDD18 pKa = 4.02LAPHH22 pKa = 6.75CFGNIPMMKK31 pKa = 8.88FCYY34 pKa = 9.79RR35 pKa = 11.84KK36 pKa = 9.81ACLRR40 pKa = 11.84LHH42 pKa = 6.79PDD44 pKa = 2.83KK45 pKa = 11.56GGDD48 pKa = 3.23AGKK51 pKa = 8.45MQRR54 pKa = 11.84LNEE57 pKa = 4.09LWQTFQQSIDD67 pKa = 3.53CLRR70 pKa = 11.84NGEE73 pKa = 4.15NAGFYY78 pKa = 10.52SFQLPNRR85 pKa = 11.84RR86 pKa = 11.84LCIWRR91 pKa = 11.84FPVFPVLLKK100 pKa = 10.47KK101 pKa = 10.56IPGASRR107 pKa = 11.84ARR109 pKa = 11.84PLLLRR114 pKa = 11.84DD115 pKa = 3.5PGFVSRR121 pKa = 11.84ISKK124 pKa = 8.91IHH126 pKa = 7.06RR127 pKa = 11.84IQHH130 pKa = 4.94HH131 pKa = 6.38HH132 pKa = 6.79CLLVSGLYY140 pKa = 10.43LPSPWKK146 pKa = 9.85EE147 pKa = 3.31ICNAFRR153 pKa = 11.84GRR155 pKa = 11.84FF156 pKa = 3.41
MM1 pKa = 8.08DD2 pKa = 4.21IALTRR7 pKa = 11.84EE8 pKa = 3.85EE9 pKa = 4.06RR10 pKa = 11.84KK11 pKa = 9.89EE12 pKa = 3.81LSEE15 pKa = 4.47LLDD18 pKa = 4.02LAPHH22 pKa = 6.75CFGNIPMMKK31 pKa = 8.88FCYY34 pKa = 9.79RR35 pKa = 11.84KK36 pKa = 9.81ACLRR40 pKa = 11.84LHH42 pKa = 6.79PDD44 pKa = 2.83KK45 pKa = 11.56GGDD48 pKa = 3.23AGKK51 pKa = 8.45MQRR54 pKa = 11.84LNEE57 pKa = 4.09LWQTFQQSIDD67 pKa = 3.53CLRR70 pKa = 11.84NGEE73 pKa = 4.15NAGFYY78 pKa = 10.52SFQLPNRR85 pKa = 11.84RR86 pKa = 11.84LCIWRR91 pKa = 11.84FPVFPVLLKK100 pKa = 10.47KK101 pKa = 10.56IPGASRR107 pKa = 11.84ARR109 pKa = 11.84PLLLRR114 pKa = 11.84DD115 pKa = 3.5PGFVSRR121 pKa = 11.84ISKK124 pKa = 8.91IHH126 pKa = 7.06RR127 pKa = 11.84IQHH130 pKa = 4.94HH131 pKa = 6.38HH132 pKa = 6.79CLLVSGLYY140 pKa = 10.43LPSPWKK146 pKa = 9.85EE147 pKa = 3.31ICNAFRR153 pKa = 11.84GRR155 pKa = 11.84FF156 pKa = 3.41
Molecular weight: 18.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2979 |
93 |
782 |
229.2 |
25.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.109 ± 0.419 | 2.685 ± 0.51 |
5.136 ± 0.33 | 5.942 ± 0.226 |
5.102 ± 0.514 | 6.613 ± 0.769 |
2.551 ± 0.383 | 4.465 ± 0.428 |
5.505 ± 0.655 | 11.715 ± 1.316 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.685 ± 0.468 | 3.39 ± 0.337 |
6.781 ± 1.036 | 4.7 ± 0.558 |
6.344 ± 0.65 | 8.493 ± 1.594 |
4.901 ± 0.573 | 3.424 ± 0.682 |
1.276 ± 0.24 | 2.182 ± 0.233 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
