
Bean leafroll virus (BLRV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Luteovirus
Average proteome isoelectric point is 7.85
Get precalculated fractions of proteins
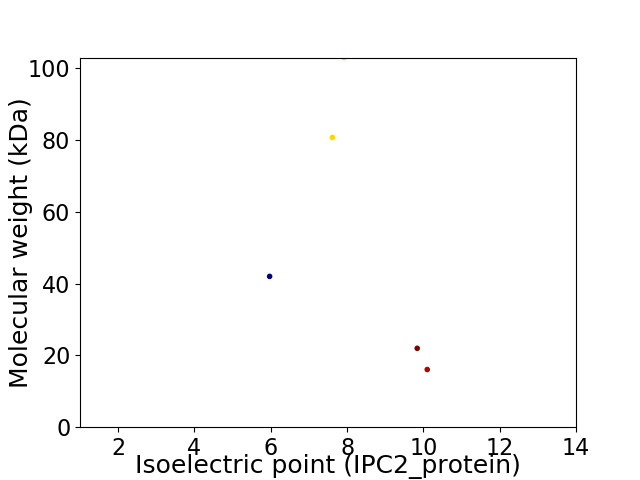
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q8V2F8|Q8V2F8_BLRV RNA dependent RNA polymerase 1 OS=Bean leafroll virus OX=12041 GN=BLRV_gp2 PE=4 SV=1
MM1 pKa = 7.47FNFDD5 pKa = 3.87SLITASSRR13 pKa = 11.84VVKK16 pKa = 10.53DD17 pKa = 5.52FIHH20 pKa = 6.26FCYY23 pKa = 10.38NRR25 pKa = 11.84ARR27 pKa = 11.84SVYY30 pKa = 9.58CALKK34 pKa = 9.25RR35 pKa = 11.84WLWEE39 pKa = 3.84LQGKK43 pKa = 9.37FDD45 pKa = 3.54AHH47 pKa = 6.06NAFVDD52 pKa = 4.01LCYY55 pKa = 10.83DD56 pKa = 3.3AMYY59 pKa = 10.46GIEE62 pKa = 4.26KK63 pKa = 10.41FEE65 pKa = 4.18EE66 pKa = 4.16EE67 pKa = 4.0LAEE70 pKa = 4.42EE71 pKa = 4.31YY72 pKa = 10.25TLAEE76 pKa = 4.24SEE78 pKa = 4.39VALAEE83 pKa = 3.75CHH85 pKa = 5.29YY86 pKa = 10.53RR87 pKa = 11.84YY88 pKa = 9.72LVSNGAPLTHH98 pKa = 6.8WPVPCPPPGAHH109 pKa = 7.13RR110 pKa = 11.84SIDD113 pKa = 3.8DD114 pKa = 3.81YY115 pKa = 11.64EE116 pKa = 5.93DD117 pKa = 3.71SVDD120 pKa = 3.37EE121 pKa = 4.55AGAFEE126 pKa = 4.3EE127 pKa = 5.11VIPSVPKK134 pKa = 10.36PEE136 pKa = 5.46PIPSKK141 pKa = 10.54DD142 pKa = 3.12GKK144 pKa = 10.38KK145 pKa = 10.17DD146 pKa = 3.4HH147 pKa = 6.91SKK149 pKa = 11.14SNPFRR154 pKa = 11.84VKK156 pKa = 10.95AFINLVKK163 pKa = 10.7AKK165 pKa = 9.87EE166 pKa = 4.02VLQATEE172 pKa = 3.79QKK174 pKa = 10.2IKK176 pKa = 10.71VVYY179 pKa = 9.72EE180 pKa = 3.99EE181 pKa = 5.15EE182 pKa = 4.05IGEE185 pKa = 4.49SPFGRR190 pKa = 11.84WFNTLPSRR198 pKa = 11.84MHH200 pKa = 5.9YY201 pKa = 9.86IKK203 pKa = 10.68RR204 pKa = 11.84ALKK207 pKa = 10.02KK208 pKa = 9.95RR209 pKa = 11.84AVNAKK214 pKa = 9.01RR215 pKa = 11.84AKK217 pKa = 10.21HH218 pKa = 4.56VTSMMKK224 pKa = 9.67EE225 pKa = 4.04VNVIPDD231 pKa = 3.94FVACCEE237 pKa = 4.3VVQVPTGEE245 pKa = 4.27MIPPKK250 pKa = 10.18KK251 pKa = 10.04NEE253 pKa = 3.95HH254 pKa = 6.31GEE256 pKa = 4.25DD257 pKa = 3.49VGEE260 pKa = 4.09PTPKK264 pKa = 9.12TKK266 pKa = 9.55MVRR269 pKa = 11.84RR270 pKa = 11.84VLPEE274 pKa = 3.59CAAEE278 pKa = 3.8ARR280 pKa = 11.84SYY282 pKa = 10.58IRR284 pKa = 11.84QHH286 pKa = 5.45IRR288 pKa = 11.84NNNMRR293 pKa = 11.84LIDD296 pKa = 4.03GTDD299 pKa = 3.23VAHH302 pKa = 6.24ATINRR307 pKa = 11.84YY308 pKa = 8.19ALKK311 pKa = 10.33FCEE314 pKa = 4.8EE315 pKa = 4.8LDD317 pKa = 3.88LDD319 pKa = 4.38LPSTTMLVDD328 pKa = 3.33YY329 pKa = 10.85AMTMVPLPLKK339 pKa = 10.75NDD341 pKa = 3.15IEE343 pKa = 4.3RR344 pKa = 11.84AKK346 pKa = 10.32IVHH349 pKa = 6.13SPNARR354 pKa = 11.84RR355 pKa = 11.84IRR357 pKa = 11.84DD358 pKa = 3.66EE359 pKa = 4.46LDD361 pKa = 3.42CLTSSVFF368 pKa = 3.37
MM1 pKa = 7.47FNFDD5 pKa = 3.87SLITASSRR13 pKa = 11.84VVKK16 pKa = 10.53DD17 pKa = 5.52FIHH20 pKa = 6.26FCYY23 pKa = 10.38NRR25 pKa = 11.84ARR27 pKa = 11.84SVYY30 pKa = 9.58CALKK34 pKa = 9.25RR35 pKa = 11.84WLWEE39 pKa = 3.84LQGKK43 pKa = 9.37FDD45 pKa = 3.54AHH47 pKa = 6.06NAFVDD52 pKa = 4.01LCYY55 pKa = 10.83DD56 pKa = 3.3AMYY59 pKa = 10.46GIEE62 pKa = 4.26KK63 pKa = 10.41FEE65 pKa = 4.18EE66 pKa = 4.16EE67 pKa = 4.0LAEE70 pKa = 4.42EE71 pKa = 4.31YY72 pKa = 10.25TLAEE76 pKa = 4.24SEE78 pKa = 4.39VALAEE83 pKa = 3.75CHH85 pKa = 5.29YY86 pKa = 10.53RR87 pKa = 11.84YY88 pKa = 9.72LVSNGAPLTHH98 pKa = 6.8WPVPCPPPGAHH109 pKa = 7.13RR110 pKa = 11.84SIDD113 pKa = 3.8DD114 pKa = 3.81YY115 pKa = 11.64EE116 pKa = 5.93DD117 pKa = 3.71SVDD120 pKa = 3.37EE121 pKa = 4.55AGAFEE126 pKa = 4.3EE127 pKa = 5.11VIPSVPKK134 pKa = 10.36PEE136 pKa = 5.46PIPSKK141 pKa = 10.54DD142 pKa = 3.12GKK144 pKa = 10.38KK145 pKa = 10.17DD146 pKa = 3.4HH147 pKa = 6.91SKK149 pKa = 11.14SNPFRR154 pKa = 11.84VKK156 pKa = 10.95AFINLVKK163 pKa = 10.7AKK165 pKa = 9.87EE166 pKa = 4.02VLQATEE172 pKa = 3.79QKK174 pKa = 10.2IKK176 pKa = 10.71VVYY179 pKa = 9.72EE180 pKa = 3.99EE181 pKa = 5.15EE182 pKa = 4.05IGEE185 pKa = 4.49SPFGRR190 pKa = 11.84WFNTLPSRR198 pKa = 11.84MHH200 pKa = 5.9YY201 pKa = 9.86IKK203 pKa = 10.68RR204 pKa = 11.84ALKK207 pKa = 10.02KK208 pKa = 9.95RR209 pKa = 11.84AVNAKK214 pKa = 9.01RR215 pKa = 11.84AKK217 pKa = 10.21HH218 pKa = 4.56VTSMMKK224 pKa = 9.67EE225 pKa = 4.04VNVIPDD231 pKa = 3.94FVACCEE237 pKa = 4.3VVQVPTGEE245 pKa = 4.27MIPPKK250 pKa = 10.18KK251 pKa = 10.04NEE253 pKa = 3.95HH254 pKa = 6.31GEE256 pKa = 4.25DD257 pKa = 3.49VGEE260 pKa = 4.09PTPKK264 pKa = 9.12TKK266 pKa = 9.55MVRR269 pKa = 11.84RR270 pKa = 11.84VLPEE274 pKa = 3.59CAAEE278 pKa = 3.8ARR280 pKa = 11.84SYY282 pKa = 10.58IRR284 pKa = 11.84QHH286 pKa = 5.45IRR288 pKa = 11.84NNNMRR293 pKa = 11.84LIDD296 pKa = 4.03GTDD299 pKa = 3.23VAHH302 pKa = 6.24ATINRR307 pKa = 11.84YY308 pKa = 8.19ALKK311 pKa = 10.33FCEE314 pKa = 4.8EE315 pKa = 4.8LDD317 pKa = 3.88LDD319 pKa = 4.38LPSTTMLVDD328 pKa = 3.33YY329 pKa = 10.85AMTMVPLPLKK339 pKa = 10.75NDD341 pKa = 3.15IEE343 pKa = 4.3RR344 pKa = 11.84AKK346 pKa = 10.32IVHH349 pKa = 6.13SPNARR354 pKa = 11.84RR355 pKa = 11.84IRR357 pKa = 11.84DD358 pKa = 3.66EE359 pKa = 4.46LDD361 pKa = 3.42CLTSSVFF368 pKa = 3.37
Molecular weight: 42.04 kDa
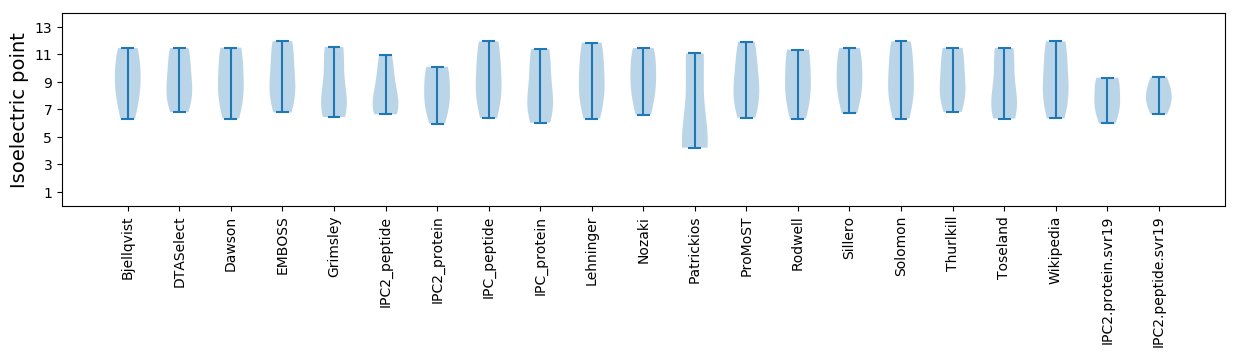
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q8V2F7|Q8V2F7_BLRV RNA-directed RNA polymerase OS=Bean leafroll virus OX=12041 PE=4 SV=1
MM1 pKa = 6.89VARR4 pKa = 11.84GKK6 pKa = 10.36RR7 pKa = 11.84VVVRR11 pKa = 11.84QLQTRR16 pKa = 11.84ARR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84LPVVLATAPVRR31 pKa = 11.84PQRR34 pKa = 11.84KK35 pKa = 8.02RR36 pKa = 11.84RR37 pKa = 11.84QRR39 pKa = 11.84GRR41 pKa = 11.84NNKK44 pKa = 9.24SRR46 pKa = 11.84GGNGFARR53 pKa = 11.84RR54 pKa = 11.84SSQVHH59 pKa = 5.03EE60 pKa = 4.61FVFSKK65 pKa = 11.2DD66 pKa = 3.54NLNGNSKK73 pKa = 10.76GSITFGPSLSEE84 pKa = 4.24CKK86 pKa = 10.04PLADD90 pKa = 5.55GILKK94 pKa = 10.25AYY96 pKa = 10.27HH97 pKa = 7.06EE98 pKa = 4.47YY99 pKa = 11.01NITNVEE105 pKa = 3.75LAYY108 pKa = 9.16ITEE111 pKa = 4.46ASSTSSGSIAYY122 pKa = 9.49EE123 pKa = 3.79LDD125 pKa = 3.12PHH127 pKa = 7.07LKK129 pKa = 8.52NTTIQSKK136 pKa = 10.11INKK139 pKa = 9.32FSITKK144 pKa = 9.48SEE146 pKa = 3.92KK147 pKa = 10.18KK148 pKa = 10.12KK149 pKa = 10.63FSRR152 pKa = 11.84KK153 pKa = 9.59AINGQAWHH161 pKa = 7.0DD162 pKa = 3.6TSEE165 pKa = 3.98DD166 pKa = 3.25QFRR169 pKa = 11.84ILYY172 pKa = 8.67EE173 pKa = 4.14GNGDD177 pKa = 3.66AKK179 pKa = 10.36IAGSFRR185 pKa = 11.84VTIKK189 pKa = 10.95VLTQNPKK196 pKa = 10.14
MM1 pKa = 6.89VARR4 pKa = 11.84GKK6 pKa = 10.36RR7 pKa = 11.84VVVRR11 pKa = 11.84QLQTRR16 pKa = 11.84ARR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84LPVVLATAPVRR31 pKa = 11.84PQRR34 pKa = 11.84KK35 pKa = 8.02RR36 pKa = 11.84RR37 pKa = 11.84QRR39 pKa = 11.84GRR41 pKa = 11.84NNKK44 pKa = 9.24SRR46 pKa = 11.84GGNGFARR53 pKa = 11.84RR54 pKa = 11.84SSQVHH59 pKa = 5.03EE60 pKa = 4.61FVFSKK65 pKa = 11.2DD66 pKa = 3.54NLNGNSKK73 pKa = 10.76GSITFGPSLSEE84 pKa = 4.24CKK86 pKa = 10.04PLADD90 pKa = 5.55GILKK94 pKa = 10.25AYY96 pKa = 10.27HH97 pKa = 7.06EE98 pKa = 4.47YY99 pKa = 11.01NITNVEE105 pKa = 3.75LAYY108 pKa = 9.16ITEE111 pKa = 4.46ASSTSSGSIAYY122 pKa = 9.49EE123 pKa = 3.79LDD125 pKa = 3.12PHH127 pKa = 7.07LKK129 pKa = 8.52NTTIQSKK136 pKa = 10.11INKK139 pKa = 9.32FSITKK144 pKa = 9.48SEE146 pKa = 3.92KK147 pKa = 10.18KK148 pKa = 10.12KK149 pKa = 10.63FSRR152 pKa = 11.84KK153 pKa = 9.59AINGQAWHH161 pKa = 7.0DD162 pKa = 3.6TSEE165 pKa = 3.98DD166 pKa = 3.25QFRR169 pKa = 11.84ILYY172 pKa = 8.67EE173 pKa = 4.14GNGDD177 pKa = 3.66AKK179 pKa = 10.36IAGSFRR185 pKa = 11.84VTIKK189 pKa = 10.95VLTQNPKK196 pKa = 10.14
Molecular weight: 21.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2329 |
142 |
901 |
465.8 |
52.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.612 ± 0.597 | 1.632 ± 0.615 |
5.195 ± 0.639 | 7.085 ± 0.763 |
3.993 ± 0.532 | 4.852 ± 0.52 |
2.319 ± 0.315 | 5.067 ± 0.359 |
7.17 ± 0.791 | 7.128 ± 0.597 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.276 ± 0.619 | 5.024 ± 0.487 |
6.483 ± 0.77 | 3.091 ± 0.541 |
7.256 ± 1.1 | 8.416 ± 1.908 |
5.067 ± 0.978 | 6.999 ± 0.933 |
1.03 ± 0.128 | 3.263 ± 0.168 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
