
Wheat dwarf India virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Mastrevirus
Average proteome isoelectric point is 7.6
Get precalculated fractions of proteins
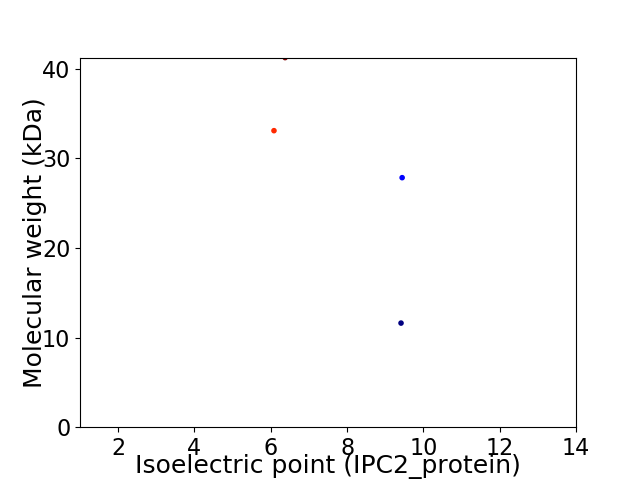
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I0E0B6|I0E0B6_9GEMI Replication-associated protein OS=Wheat dwarf India virus OX=1174526 GN=RepA PE=3 SV=1
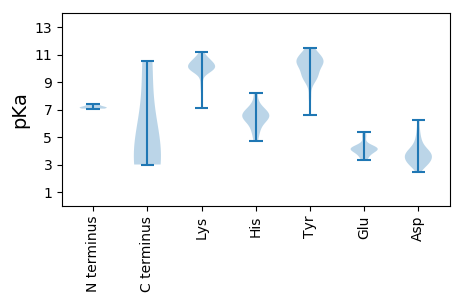
MM1 pKa = 7.16SQTSAEE7 pKa = 4.15NNSANPKK14 pKa = 9.85ASSSTFRR21 pKa = 11.84YY22 pKa = 9.66RR23 pKa = 11.84SNNCFLTFPHH33 pKa = 6.85CNSCPYY39 pKa = 11.32GMVQHH44 pKa = 6.51FWDD47 pKa = 6.2LISTWSPIYY56 pKa = 10.52AVASVEE62 pKa = 3.96LHH64 pKa = 6.31QDD66 pKa = 3.6GTPHH70 pKa = 7.04LHH72 pKa = 7.39ALLQTRR78 pKa = 11.84KK79 pKa = 9.99QISTNDD85 pKa = 3.09PHH87 pKa = 8.17FFDD90 pKa = 5.45FDD92 pKa = 3.48GHH94 pKa = 6.39HH95 pKa = 6.67PHH97 pKa = 6.45IQAAKK102 pKa = 10.2NPTLCRR108 pKa = 11.84DD109 pKa = 4.1YY110 pKa = 10.96ILKK113 pKa = 10.76GPITFSEE120 pKa = 4.21KK121 pKa = 10.4GAFIPRR127 pKa = 11.84GRR129 pKa = 11.84NAGTSPRR136 pKa = 11.84HH137 pKa = 4.7SNKK140 pKa = 9.68RR141 pKa = 11.84SRR143 pKa = 11.84DD144 pKa = 3.83DD145 pKa = 3.16IMKK148 pKa = 10.33DD149 pKa = 2.87IIEE152 pKa = 4.36NSTNKK157 pKa = 9.72SDD159 pKa = 3.82YY160 pKa = 10.04LSKK163 pKa = 10.55VRR165 pKa = 11.84RR166 pKa = 11.84NFPYY170 pKa = 10.46DD171 pKa = 3.17WATKK175 pKa = 10.05LYY177 pKa = 9.47NFEE180 pKa = 4.19YY181 pKa = 10.26SASKK185 pKa = 10.41LFPEE189 pKa = 4.14QQPEE193 pKa = 3.89YY194 pKa = 10.27SNPHH198 pKa = 5.63GQSVPDD204 pKa = 3.91LYY206 pKa = 11.03CYY208 pKa = 7.88EE209 pKa = 5.18TIQDD213 pKa = 4.18WIDD216 pKa = 3.54SNLFQVHH223 pKa = 6.81PYY225 pKa = 10.52CYY227 pKa = 9.48MLNTPTINSVAEE239 pKa = 4.03AVQEE243 pKa = 4.11LTWLQEE249 pKa = 3.57NSRR252 pKa = 11.84IHH254 pKa = 6.19LQEE257 pKa = 3.56QGQRR261 pKa = 11.84ASTSSVQQGPEE272 pKa = 3.86NLHH275 pKa = 6.18GQGPWADD282 pKa = 3.62TTTGRR287 pKa = 11.84IMM289 pKa = 4.33
MM1 pKa = 7.16SQTSAEE7 pKa = 4.15NNSANPKK14 pKa = 9.85ASSSTFRR21 pKa = 11.84YY22 pKa = 9.66RR23 pKa = 11.84SNNCFLTFPHH33 pKa = 6.85CNSCPYY39 pKa = 11.32GMVQHH44 pKa = 6.51FWDD47 pKa = 6.2LISTWSPIYY56 pKa = 10.52AVASVEE62 pKa = 3.96LHH64 pKa = 6.31QDD66 pKa = 3.6GTPHH70 pKa = 7.04LHH72 pKa = 7.39ALLQTRR78 pKa = 11.84KK79 pKa = 9.99QISTNDD85 pKa = 3.09PHH87 pKa = 8.17FFDD90 pKa = 5.45FDD92 pKa = 3.48GHH94 pKa = 6.39HH95 pKa = 6.67PHH97 pKa = 6.45IQAAKK102 pKa = 10.2NPTLCRR108 pKa = 11.84DD109 pKa = 4.1YY110 pKa = 10.96ILKK113 pKa = 10.76GPITFSEE120 pKa = 4.21KK121 pKa = 10.4GAFIPRR127 pKa = 11.84GRR129 pKa = 11.84NAGTSPRR136 pKa = 11.84HH137 pKa = 4.7SNKK140 pKa = 9.68RR141 pKa = 11.84SRR143 pKa = 11.84DD144 pKa = 3.83DD145 pKa = 3.16IMKK148 pKa = 10.33DD149 pKa = 2.87IIEE152 pKa = 4.36NSTNKK157 pKa = 9.72SDD159 pKa = 3.82YY160 pKa = 10.04LSKK163 pKa = 10.55VRR165 pKa = 11.84RR166 pKa = 11.84NFPYY170 pKa = 10.46DD171 pKa = 3.17WATKK175 pKa = 10.05LYY177 pKa = 9.47NFEE180 pKa = 4.19YY181 pKa = 10.26SASKK185 pKa = 10.41LFPEE189 pKa = 4.14QQPEE193 pKa = 3.89YY194 pKa = 10.27SNPHH198 pKa = 5.63GQSVPDD204 pKa = 3.91LYY206 pKa = 11.03CYY208 pKa = 7.88EE209 pKa = 5.18TIQDD213 pKa = 4.18WIDD216 pKa = 3.54SNLFQVHH223 pKa = 6.81PYY225 pKa = 10.52CYY227 pKa = 9.48MLNTPTINSVAEE239 pKa = 4.03AVQEE243 pKa = 4.11LTWLQEE249 pKa = 3.57NSRR252 pKa = 11.84IHH254 pKa = 6.19LQEE257 pKa = 3.56QGQRR261 pKa = 11.84ASTSSVQQGPEE272 pKa = 3.86NLHH275 pKa = 6.18GQGPWADD282 pKa = 3.62TTTGRR287 pKa = 11.84IMM289 pKa = 4.33
Molecular weight: 33.08 kDa
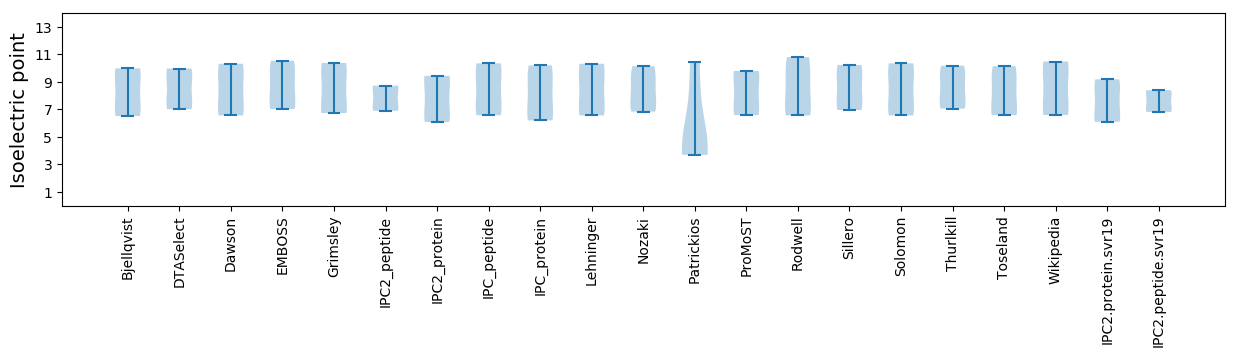
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I0E0B3|I0E0B3_9GEMI Movement protein OS=Wheat dwarf India virus OX=1174526 GN=V1 PE=3 SV=1
MM1 pKa = 7.06SQVKK5 pKa = 9.91KK6 pKa = 8.96RR7 pKa = 11.84TRR9 pKa = 11.84RR10 pKa = 11.84GDD12 pKa = 3.32EE13 pKa = 4.14VEE15 pKa = 3.38WSKK18 pKa = 11.15RR19 pKa = 11.84KK20 pKa = 9.64RR21 pKa = 11.84LRR23 pKa = 11.84SSGSKK28 pKa = 10.74GSATKK33 pKa = 10.25LRR35 pKa = 11.84KK36 pKa = 9.92LPVRR40 pKa = 11.84PSLQIQSLYY49 pKa = 9.24YY50 pKa = 10.43TLTTYY55 pKa = 11.11VDD57 pKa = 4.65FKK59 pKa = 11.03TGGEE63 pKa = 4.27SYY65 pKa = 11.47LMASYY70 pKa = 11.26ARR72 pKa = 11.84GSDD75 pKa = 2.92EE76 pKa = 4.93GEE78 pKa = 3.7RR79 pKa = 11.84HH80 pKa = 5.31TNEE83 pKa = 3.34TMTYY87 pKa = 9.1KK88 pKa = 10.78AALNLHH94 pKa = 6.77LSASSDD100 pKa = 2.91ACKK103 pKa = 10.58YY104 pKa = 10.45SNTGTGCIWLIYY116 pKa = 9.59DD117 pKa = 4.79AQPTGNKK124 pKa = 9.14PSPKK128 pKa = 10.14DD129 pKa = 2.95IFGYY133 pKa = 10.22SDD135 pKa = 4.65SLNSWPTTWMVSRR148 pKa = 11.84EE149 pKa = 3.87QCHH152 pKa = 5.29RR153 pKa = 11.84FVVKK157 pKa = 10.48RR158 pKa = 11.84RR159 pKa = 11.84WTFTLEE165 pKa = 4.16TNGRR169 pKa = 11.84LASDD173 pKa = 4.03VPPSNVVWPPCKK185 pKa = 10.05KK186 pKa = 10.47SINFHH191 pKa = 6.3KK192 pKa = 9.94FAKK195 pKa = 9.91GLGVRR200 pKa = 11.84TEE202 pKa = 4.24WKK204 pKa = 10.07NKK206 pKa = 7.1TDD208 pKa = 3.57GGIGSIKK215 pKa = 10.24KK216 pKa = 8.67GALYY220 pKa = 9.5FVIAPGNGVEE230 pKa = 4.03GRR232 pKa = 11.84MFGTIRR238 pKa = 11.84MYY240 pKa = 10.66FKK242 pKa = 11.0SIGNQQ247 pKa = 3.0
MM1 pKa = 7.06SQVKK5 pKa = 9.91KK6 pKa = 8.96RR7 pKa = 11.84TRR9 pKa = 11.84RR10 pKa = 11.84GDD12 pKa = 3.32EE13 pKa = 4.14VEE15 pKa = 3.38WSKK18 pKa = 11.15RR19 pKa = 11.84KK20 pKa = 9.64RR21 pKa = 11.84LRR23 pKa = 11.84SSGSKK28 pKa = 10.74GSATKK33 pKa = 10.25LRR35 pKa = 11.84KK36 pKa = 9.92LPVRR40 pKa = 11.84PSLQIQSLYY49 pKa = 9.24YY50 pKa = 10.43TLTTYY55 pKa = 11.11VDD57 pKa = 4.65FKK59 pKa = 11.03TGGEE63 pKa = 4.27SYY65 pKa = 11.47LMASYY70 pKa = 11.26ARR72 pKa = 11.84GSDD75 pKa = 2.92EE76 pKa = 4.93GEE78 pKa = 3.7RR79 pKa = 11.84HH80 pKa = 5.31TNEE83 pKa = 3.34TMTYY87 pKa = 9.1KK88 pKa = 10.78AALNLHH94 pKa = 6.77LSASSDD100 pKa = 2.91ACKK103 pKa = 10.58YY104 pKa = 10.45SNTGTGCIWLIYY116 pKa = 9.59DD117 pKa = 4.79AQPTGNKK124 pKa = 9.14PSPKK128 pKa = 10.14DD129 pKa = 2.95IFGYY133 pKa = 10.22SDD135 pKa = 4.65SLNSWPTTWMVSRR148 pKa = 11.84EE149 pKa = 3.87QCHH152 pKa = 5.29RR153 pKa = 11.84FVVKK157 pKa = 10.48RR158 pKa = 11.84RR159 pKa = 11.84WTFTLEE165 pKa = 4.16TNGRR169 pKa = 11.84LASDD173 pKa = 4.03VPPSNVVWPPCKK185 pKa = 10.05KK186 pKa = 10.47SINFHH191 pKa = 6.3KK192 pKa = 9.94FAKK195 pKa = 9.91GLGVRR200 pKa = 11.84TEE202 pKa = 4.24WKK204 pKa = 10.07NKK206 pKa = 7.1TDD208 pKa = 3.57GGIGSIKK215 pKa = 10.24KK216 pKa = 8.67GALYY220 pKa = 9.5FVIAPGNGVEE230 pKa = 4.03GRR232 pKa = 11.84MFGTIRR238 pKa = 11.84MYY240 pKa = 10.66FKK242 pKa = 11.0SIGNQQ247 pKa = 3.0
Molecular weight: 27.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
999 |
106 |
357 |
249.8 |
28.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.906 ± 0.582 | 1.802 ± 0.232 |
4.905 ± 0.445 | 4.304 ± 0.109 |
4.705 ± 0.393 | 6.106 ± 1.017 |
3.103 ± 0.693 | 5.606 ± 0.514 |
5.506 ± 0.977 | 6.106 ± 0.301 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.902 ± 0.152 | 5.706 ± 0.654 |
7.007 ± 0.754 | 4.505 ± 0.763 |
5.706 ± 0.728 | 9.109 ± 0.532 |
6.607 ± 0.499 | 4.204 ± 0.486 |
2.302 ± 0.139 | 4.905 ± 0.289 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
