
Paramicrosporidium saccamoebae
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Fungi incertae sedis; Cryptomycota; Cryptomycota incertae sedis; Paramicrosporidium
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3754 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
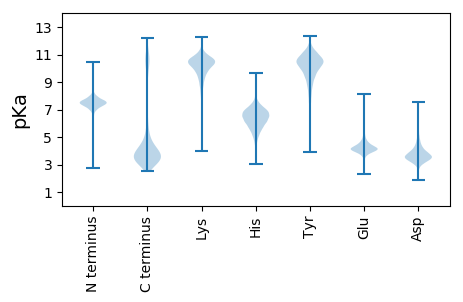
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A2H9TMI5|A0A2H9TMI5_9FUNG Putative serine threonine protein kinase OS=Paramicrosporidium saccamoebae OX=1246581 GN=PSACC_01238 PE=3 SV=1
MM1 pKa = 7.61SPPGLIRR8 pKa = 11.84IKK10 pKa = 10.49RR11 pKa = 11.84SAEE14 pKa = 3.98DD15 pKa = 3.78PPLVALLCEE24 pKa = 4.35HH25 pKa = 7.57DD26 pKa = 5.11EE27 pKa = 4.28DD28 pKa = 3.99TLYY31 pKa = 10.52YY32 pKa = 10.83QLMHH36 pKa = 6.96HH37 pKa = 7.08DD38 pKa = 4.33EE39 pKa = 4.5SNKK42 pKa = 7.57TACVEE47 pKa = 3.89PVTLSEE53 pKa = 4.23ALEE56 pKa = 4.1QIKK59 pKa = 10.38IEE61 pKa = 4.06ALPVKK66 pKa = 9.55MDD68 pKa = 3.57EE69 pKa = 4.52VKK71 pKa = 11.13LEE73 pKa = 4.31DD74 pKa = 3.47EE75 pKa = 4.36TLDD78 pKa = 3.55GFVYY82 pKa = 10.33DD83 pKa = 4.16YY84 pKa = 11.18YY85 pKa = 10.83LHH87 pKa = 7.25KK88 pKa = 10.43STAPEE93 pKa = 3.7QLDD96 pKa = 3.39VSRR99 pKa = 11.84IDD101 pKa = 4.13EE102 pKa = 4.5EE103 pKa = 4.94DD104 pKa = 3.79SLWEE108 pKa = 4.92DD109 pKa = 4.8DD110 pKa = 4.07EE111 pKa = 5.73DD112 pKa = 5.75SNAEE116 pKa = 4.07DD117 pKa = 5.19YY118 pKa = 10.44YY119 pKa = 11.53QNDD122 pKa = 3.86YY123 pKa = 10.95PDD125 pKa = 4.6EE126 pKa = 6.13DD127 pKa = 5.25DD128 pKa = 5.55DD129 pKa = 5.64NSDD132 pKa = 3.37VTSNEE137 pKa = 4.83DD138 pKa = 2.83SDD140 pKa = 2.96HH141 pKa = 6.81WYY143 pKa = 10.39RR144 pKa = 11.84SGLSRR149 pKa = 11.84HH150 pKa = 6.48HH151 pKa = 7.26DD152 pKa = 3.5GCNSGFSDD160 pKa = 4.38DD161 pKa = 4.64DD162 pKa = 4.32AYY164 pKa = 11.12
MM1 pKa = 7.61SPPGLIRR8 pKa = 11.84IKK10 pKa = 10.49RR11 pKa = 11.84SAEE14 pKa = 3.98DD15 pKa = 3.78PPLVALLCEE24 pKa = 4.35HH25 pKa = 7.57DD26 pKa = 5.11EE27 pKa = 4.28DD28 pKa = 3.99TLYY31 pKa = 10.52YY32 pKa = 10.83QLMHH36 pKa = 6.96HH37 pKa = 7.08DD38 pKa = 4.33EE39 pKa = 4.5SNKK42 pKa = 7.57TACVEE47 pKa = 3.89PVTLSEE53 pKa = 4.23ALEE56 pKa = 4.1QIKK59 pKa = 10.38IEE61 pKa = 4.06ALPVKK66 pKa = 9.55MDD68 pKa = 3.57EE69 pKa = 4.52VKK71 pKa = 11.13LEE73 pKa = 4.31DD74 pKa = 3.47EE75 pKa = 4.36TLDD78 pKa = 3.55GFVYY82 pKa = 10.33DD83 pKa = 4.16YY84 pKa = 11.18YY85 pKa = 10.83LHH87 pKa = 7.25KK88 pKa = 10.43STAPEE93 pKa = 3.7QLDD96 pKa = 3.39VSRR99 pKa = 11.84IDD101 pKa = 4.13EE102 pKa = 4.5EE103 pKa = 4.94DD104 pKa = 3.79SLWEE108 pKa = 4.92DD109 pKa = 4.8DD110 pKa = 4.07EE111 pKa = 5.73DD112 pKa = 5.75SNAEE116 pKa = 4.07DD117 pKa = 5.19YY118 pKa = 10.44YY119 pKa = 11.53QNDD122 pKa = 3.86YY123 pKa = 10.95PDD125 pKa = 4.6EE126 pKa = 6.13DD127 pKa = 5.25DD128 pKa = 5.55DD129 pKa = 5.64NSDD132 pKa = 3.37VTSNEE137 pKa = 4.83DD138 pKa = 2.83SDD140 pKa = 2.96HH141 pKa = 6.81WYY143 pKa = 10.39RR144 pKa = 11.84SGLSRR149 pKa = 11.84HH150 pKa = 6.48HH151 pKa = 7.26DD152 pKa = 3.5GCNSGFSDD160 pKa = 4.38DD161 pKa = 4.64DD162 pKa = 4.32AYY164 pKa = 11.12
Molecular weight: 18.89 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H9TJL1|A0A2H9TJL1_9FUNG Mitochondrial phosphate carrier protein 2 OS=Paramicrosporidium saccamoebae OX=1246581 GN=PSACC_02238 PE=3 SV=1
MM1 pKa = 7.53HH2 pKa = 7.75LNTFIIASIVPMALGFMSIPNRR24 pKa = 11.84WVRR27 pKa = 11.84HH28 pKa = 4.85CRR30 pKa = 11.84AGHH33 pKa = 4.81TRR35 pKa = 11.84AIRR38 pKa = 11.84LADD41 pKa = 3.94CDD43 pKa = 3.82EE44 pKa = 4.85LGTRR48 pKa = 11.84GMEE51 pKa = 3.78QCLMSTHH58 pKa = 6.91DD59 pKa = 3.51RR60 pKa = 11.84TLAALIRR67 pKa = 11.84RR68 pKa = 11.84CPNQLPKK75 pKa = 10.49FRR77 pKa = 11.84DD78 pKa = 3.38FDD80 pKa = 3.95RR81 pKa = 11.84FALRR85 pKa = 11.84LYY87 pKa = 10.75QSFLPRR93 pKa = 11.84AFRR96 pKa = 11.84AYY98 pKa = 9.32IQTRR102 pKa = 11.84CGQLSRR108 pKa = 11.84STLRR112 pKa = 11.84EE113 pKa = 3.83LLNSFVNNNSVPFVRR128 pKa = 11.84AVMQHH133 pKa = 5.59CVVGEE138 pKa = 4.1RR139 pKa = 11.84IWRR142 pKa = 11.84QFAQQARR149 pKa = 11.84LQGKK153 pKa = 9.19NEE155 pKa = 4.02LLVVLRR161 pKa = 11.84AGPAGPVGQPGQSGQSGQSGQSGQLGQGAVQDD193 pKa = 4.17DD194 pKa = 4.66SKK196 pKa = 11.63NSWGMSYY203 pKa = 10.28SVKK206 pKa = 10.19RR207 pKa = 11.84DD208 pKa = 3.36VHH210 pKa = 5.4TMPSGEE216 pKa = 4.02RR217 pKa = 11.84FAVARR222 pKa = 11.84QEE224 pKa = 3.77ASKK227 pKa = 11.14DD228 pKa = 3.4GGIQPGALGQNGGLAAGTLGQSGLADD254 pKa = 3.5PTFGGSAQVNQQRR267 pKa = 11.84ATSKK271 pKa = 10.07RR272 pKa = 11.84AYY274 pKa = 10.63LKK276 pKa = 9.67VTTQQVASNLNILGRR291 pKa = 11.84LTKK294 pKa = 10.51AQASTFGMLSDD305 pKa = 4.19GCSALTEE312 pKa = 4.18SHH314 pKa = 6.76FRR316 pKa = 11.84QPTVSPWIVRR326 pKa = 11.84SLPQEE331 pKa = 4.03CFVRR335 pKa = 11.84IPPPAFSGLTPAMISQIKK353 pKa = 7.97WWPFVTRR360 pKa = 11.84DD361 pKa = 3.36QIRR364 pKa = 11.84YY365 pKa = 9.18VPTGDD370 pKa = 4.93AIRR373 pKa = 11.84ALPFDD378 pKa = 3.51QLGKK382 pKa = 10.59GRR384 pKa = 11.84QKK386 pKa = 10.9DD387 pKa = 3.88GEE389 pKa = 4.3DD390 pKa = 3.5RR391 pKa = 11.84EE392 pKa = 4.72HH393 pKa = 6.9PCWTITRR400 pKa = 11.84DD401 pKa = 3.26QMRR404 pKa = 11.84SIRR407 pKa = 11.84RR408 pKa = 11.84SKK410 pKa = 9.73TVFKK414 pKa = 10.01EE415 pKa = 3.57YY416 pKa = 10.65SRR418 pKa = 11.84RR419 pKa = 11.84CVKK422 pKa = 10.51SGASAVVVSMSSDD435 pKa = 3.18FRR437 pKa = 11.84FPLGGPPLHH446 pKa = 6.53KK447 pKa = 10.14QRR449 pKa = 11.84PNQYY453 pKa = 10.44ARR455 pKa = 11.84AEE457 pKa = 4.38DD458 pKa = 3.98III460 pKa = 5.03
MM1 pKa = 7.53HH2 pKa = 7.75LNTFIIASIVPMALGFMSIPNRR24 pKa = 11.84WVRR27 pKa = 11.84HH28 pKa = 4.85CRR30 pKa = 11.84AGHH33 pKa = 4.81TRR35 pKa = 11.84AIRR38 pKa = 11.84LADD41 pKa = 3.94CDD43 pKa = 3.82EE44 pKa = 4.85LGTRR48 pKa = 11.84GMEE51 pKa = 3.78QCLMSTHH58 pKa = 6.91DD59 pKa = 3.51RR60 pKa = 11.84TLAALIRR67 pKa = 11.84RR68 pKa = 11.84CPNQLPKK75 pKa = 10.49FRR77 pKa = 11.84DD78 pKa = 3.38FDD80 pKa = 3.95RR81 pKa = 11.84FALRR85 pKa = 11.84LYY87 pKa = 10.75QSFLPRR93 pKa = 11.84AFRR96 pKa = 11.84AYY98 pKa = 9.32IQTRR102 pKa = 11.84CGQLSRR108 pKa = 11.84STLRR112 pKa = 11.84EE113 pKa = 3.83LLNSFVNNNSVPFVRR128 pKa = 11.84AVMQHH133 pKa = 5.59CVVGEE138 pKa = 4.1RR139 pKa = 11.84IWRR142 pKa = 11.84QFAQQARR149 pKa = 11.84LQGKK153 pKa = 9.19NEE155 pKa = 4.02LLVVLRR161 pKa = 11.84AGPAGPVGQPGQSGQSGQSGQSGQLGQGAVQDD193 pKa = 4.17DD194 pKa = 4.66SKK196 pKa = 11.63NSWGMSYY203 pKa = 10.28SVKK206 pKa = 10.19RR207 pKa = 11.84DD208 pKa = 3.36VHH210 pKa = 5.4TMPSGEE216 pKa = 4.02RR217 pKa = 11.84FAVARR222 pKa = 11.84QEE224 pKa = 3.77ASKK227 pKa = 11.14DD228 pKa = 3.4GGIQPGALGQNGGLAAGTLGQSGLADD254 pKa = 3.5PTFGGSAQVNQQRR267 pKa = 11.84ATSKK271 pKa = 10.07RR272 pKa = 11.84AYY274 pKa = 10.63LKK276 pKa = 9.67VTTQQVASNLNILGRR291 pKa = 11.84LTKK294 pKa = 10.51AQASTFGMLSDD305 pKa = 4.19GCSALTEE312 pKa = 4.18SHH314 pKa = 6.76FRR316 pKa = 11.84QPTVSPWIVRR326 pKa = 11.84SLPQEE331 pKa = 4.03CFVRR335 pKa = 11.84IPPPAFSGLTPAMISQIKK353 pKa = 7.97WWPFVTRR360 pKa = 11.84DD361 pKa = 3.36QIRR364 pKa = 11.84YY365 pKa = 9.18VPTGDD370 pKa = 4.93AIRR373 pKa = 11.84ALPFDD378 pKa = 3.51QLGKK382 pKa = 10.59GRR384 pKa = 11.84QKK386 pKa = 10.9DD387 pKa = 3.88GEE389 pKa = 4.3DD390 pKa = 3.5RR391 pKa = 11.84EE392 pKa = 4.72HH393 pKa = 6.9PCWTITRR400 pKa = 11.84DD401 pKa = 3.26QMRR404 pKa = 11.84SIRR407 pKa = 11.84RR408 pKa = 11.84SKK410 pKa = 9.73TVFKK414 pKa = 10.01EE415 pKa = 3.57YY416 pKa = 10.65SRR418 pKa = 11.84RR419 pKa = 11.84CVKK422 pKa = 10.51SGASAVVVSMSSDD435 pKa = 3.18FRR437 pKa = 11.84FPLGGPPLHH446 pKa = 6.53KK447 pKa = 10.14QRR449 pKa = 11.84PNQYY453 pKa = 10.44ARR455 pKa = 11.84AEE457 pKa = 4.38DD458 pKa = 3.98III460 pKa = 5.03
Molecular weight: 50.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1669180 |
36 |
6083 |
444.6 |
49.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.805 ± 0.034 | 1.603 ± 0.016 |
5.41 ± 0.025 | 6.509 ± 0.037 |
3.965 ± 0.025 | 5.785 ± 0.035 |
2.352 ± 0.017 | 5.411 ± 0.026 |
5.222 ± 0.043 | 10.355 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.531 ± 0.016 | 3.629 ± 0.018 |
5.066 ± 0.032 | 3.807 ± 0.022 |
6.146 ± 0.034 | 7.886 ± 0.041 |
5.76 ± 0.039 | 6.718 ± 0.028 |
1.24 ± 0.013 | 2.776 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
