
Tortoise microvirus 48
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
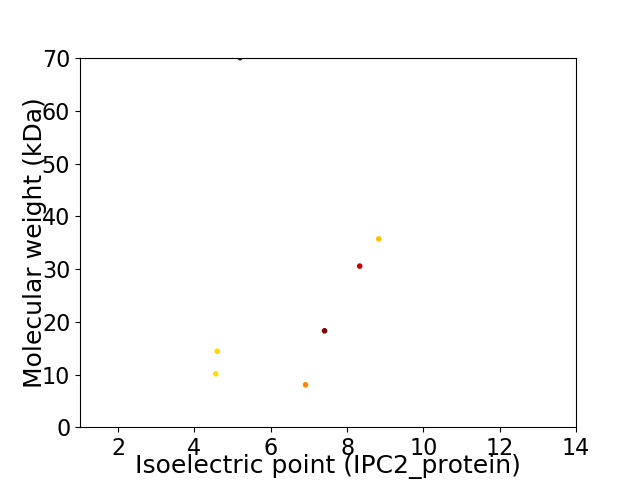
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
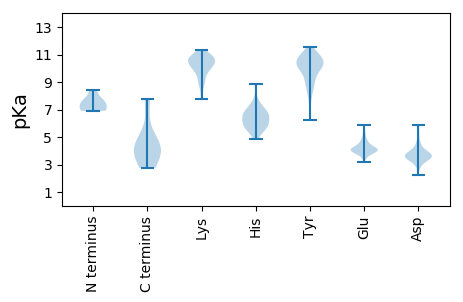
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A4P8W9P0|A0A4P8W9P0_9VIRU Uncharacterized protein OS=Tortoise microvirus 48 OX=2583152 PE=4 SV=1
MM1 pKa = 6.9KK2 pKa = 9.93TNVYY6 pKa = 10.43SIIDD10 pKa = 3.67SAYY13 pKa = 9.78HH14 pKa = 5.78RR15 pKa = 11.84VQGIVVAPNSSLAIRR30 pKa = 11.84DD31 pKa = 3.92VAPMLQRR38 pKa = 11.84VAPHH42 pKa = 6.47FEE44 pKa = 3.37EE45 pKa = 5.9DD46 pKa = 3.83MYY48 pKa = 11.12IEE50 pKa = 4.83CIGYY54 pKa = 10.2YY55 pKa = 10.45DD56 pKa = 5.45DD57 pKa = 5.91DD58 pKa = 5.21LSFHH62 pKa = 5.36VTSPEE67 pKa = 4.04VFPWTADD74 pKa = 3.48NPEE77 pKa = 4.2VPMKK81 pKa = 10.82ALTGKK86 pKa = 8.94DD87 pKa = 3.31TVII90 pKa = 4.35
MM1 pKa = 6.9KK2 pKa = 9.93TNVYY6 pKa = 10.43SIIDD10 pKa = 3.67SAYY13 pKa = 9.78HH14 pKa = 5.78RR15 pKa = 11.84VQGIVVAPNSSLAIRR30 pKa = 11.84DD31 pKa = 3.92VAPMLQRR38 pKa = 11.84VAPHH42 pKa = 6.47FEE44 pKa = 3.37EE45 pKa = 5.9DD46 pKa = 3.83MYY48 pKa = 11.12IEE50 pKa = 4.83CIGYY54 pKa = 10.2YY55 pKa = 10.45DD56 pKa = 5.45DD57 pKa = 5.91DD58 pKa = 5.21LSFHH62 pKa = 5.36VTSPEE67 pKa = 4.04VFPWTADD74 pKa = 3.48NPEE77 pKa = 4.2VPMKK81 pKa = 10.82ALTGKK86 pKa = 8.94DD87 pKa = 3.31TVII90 pKa = 4.35
Molecular weight: 10.13 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W6M6|A0A4P8W6M6_9VIRU Uncharacterized protein OS=Tortoise microvirus 48 OX=2583152 PE=4 SV=1
MM1 pKa = 7.48ACSSPFFLEE10 pKa = 4.06TVKK13 pKa = 10.55DD14 pKa = 3.71YY15 pKa = 11.08RR16 pKa = 11.84GRR18 pKa = 11.84PVPLACRR25 pKa = 11.84VCNTCRR31 pKa = 11.84ADD33 pKa = 3.82RR34 pKa = 11.84VSYY37 pKa = 8.76WEE39 pKa = 4.1RR40 pKa = 11.84RR41 pKa = 11.84AKK43 pKa = 10.82YY44 pKa = 8.62EE45 pKa = 3.93WNNSISSAFVTFTYY59 pKa = 10.93DD60 pKa = 3.19NDD62 pKa = 3.46NVPIGRR68 pKa = 11.84TGQQSLRR75 pKa = 11.84RR76 pKa = 11.84DD77 pKa = 3.55DD78 pKa = 3.4LHH80 pKa = 8.88RR81 pKa = 11.84YY82 pKa = 9.35IDD84 pKa = 3.83TLRR87 pKa = 11.84HH88 pKa = 5.71RR89 pKa = 11.84LKK91 pKa = 10.44KK92 pKa = 10.73DD93 pKa = 2.78KK94 pKa = 10.64VLPARR99 pKa = 11.84CIPNFSFIACGEE111 pKa = 4.18YY112 pKa = 10.44GDD114 pKa = 4.57SFGRR118 pKa = 11.84PHH120 pKa = 5.56YY121 pKa = 10.23HH122 pKa = 5.88VLFFGLDD129 pKa = 3.43FANCAKK135 pKa = 10.21YY136 pKa = 10.83LKK138 pKa = 9.46TSWHH142 pKa = 5.97NGSIKK147 pKa = 10.15VDD149 pKa = 3.64PVKK152 pKa = 10.94NGAIRR157 pKa = 11.84YY158 pKa = 6.96VISYY162 pKa = 8.12MSKK165 pKa = 9.76EE166 pKa = 4.12LYY168 pKa = 10.05GDD170 pKa = 3.65RR171 pKa = 11.84RR172 pKa = 11.84DD173 pKa = 3.59EE174 pKa = 5.44EE175 pKa = 4.46YY176 pKa = 10.95FDD178 pKa = 4.01WDD180 pKa = 3.11IEE182 pKa = 4.5APFLMCSRR190 pKa = 11.84GFGSGLFEE198 pKa = 4.06SQRR201 pKa = 11.84NNIRR205 pKa = 11.84EE206 pKa = 4.02YY207 pKa = 11.08GAMKK211 pKa = 10.47FGTKK215 pKa = 9.98FFSVPPYY222 pKa = 9.36WKK224 pKa = 10.62NKK226 pKa = 9.26FIDD229 pKa = 3.59MDD231 pKa = 3.72IEE233 pKa = 4.19KK234 pKa = 10.8ADD236 pKa = 3.43ARR238 pKa = 11.84YY239 pKa = 10.0EE240 pKa = 3.88NARR243 pKa = 11.84RR244 pKa = 11.84IQFNNWSANGGHH256 pKa = 6.57FGSPYY261 pKa = 8.88KK262 pKa = 10.88AQMQKK267 pKa = 8.9TKK269 pKa = 10.21IRR271 pKa = 11.84AEE273 pKa = 4.07SLYY276 pKa = 10.52HH277 pKa = 6.67DD278 pKa = 3.65MLMRR282 pKa = 11.84GSKK285 pKa = 9.49IGYY288 pKa = 7.92KK289 pKa = 9.98RR290 pKa = 11.84PVYY293 pKa = 9.67MYY295 pKa = 9.99GGSGSKK301 pKa = 10.4LVGLIEE307 pKa = 4.04
MM1 pKa = 7.48ACSSPFFLEE10 pKa = 4.06TVKK13 pKa = 10.55DD14 pKa = 3.71YY15 pKa = 11.08RR16 pKa = 11.84GRR18 pKa = 11.84PVPLACRR25 pKa = 11.84VCNTCRR31 pKa = 11.84ADD33 pKa = 3.82RR34 pKa = 11.84VSYY37 pKa = 8.76WEE39 pKa = 4.1RR40 pKa = 11.84RR41 pKa = 11.84AKK43 pKa = 10.82YY44 pKa = 8.62EE45 pKa = 3.93WNNSISSAFVTFTYY59 pKa = 10.93DD60 pKa = 3.19NDD62 pKa = 3.46NVPIGRR68 pKa = 11.84TGQQSLRR75 pKa = 11.84RR76 pKa = 11.84DD77 pKa = 3.55DD78 pKa = 3.4LHH80 pKa = 8.88RR81 pKa = 11.84YY82 pKa = 9.35IDD84 pKa = 3.83TLRR87 pKa = 11.84HH88 pKa = 5.71RR89 pKa = 11.84LKK91 pKa = 10.44KK92 pKa = 10.73DD93 pKa = 2.78KK94 pKa = 10.64VLPARR99 pKa = 11.84CIPNFSFIACGEE111 pKa = 4.18YY112 pKa = 10.44GDD114 pKa = 4.57SFGRR118 pKa = 11.84PHH120 pKa = 5.56YY121 pKa = 10.23HH122 pKa = 5.88VLFFGLDD129 pKa = 3.43FANCAKK135 pKa = 10.21YY136 pKa = 10.83LKK138 pKa = 9.46TSWHH142 pKa = 5.97NGSIKK147 pKa = 10.15VDD149 pKa = 3.64PVKK152 pKa = 10.94NGAIRR157 pKa = 11.84YY158 pKa = 6.96VISYY162 pKa = 8.12MSKK165 pKa = 9.76EE166 pKa = 4.12LYY168 pKa = 10.05GDD170 pKa = 3.65RR171 pKa = 11.84RR172 pKa = 11.84DD173 pKa = 3.59EE174 pKa = 5.44EE175 pKa = 4.46YY176 pKa = 10.95FDD178 pKa = 4.01WDD180 pKa = 3.11IEE182 pKa = 4.5APFLMCSRR190 pKa = 11.84GFGSGLFEE198 pKa = 4.06SQRR201 pKa = 11.84NNIRR205 pKa = 11.84EE206 pKa = 4.02YY207 pKa = 11.08GAMKK211 pKa = 10.47FGTKK215 pKa = 9.98FFSVPPYY222 pKa = 9.36WKK224 pKa = 10.62NKK226 pKa = 9.26FIDD229 pKa = 3.59MDD231 pKa = 3.72IEE233 pKa = 4.19KK234 pKa = 10.8ADD236 pKa = 3.43ARR238 pKa = 11.84YY239 pKa = 10.0EE240 pKa = 3.88NARR243 pKa = 11.84RR244 pKa = 11.84IQFNNWSANGGHH256 pKa = 6.57FGSPYY261 pKa = 8.88KK262 pKa = 10.88AQMQKK267 pKa = 8.9TKK269 pKa = 10.21IRR271 pKa = 11.84AEE273 pKa = 4.07SLYY276 pKa = 10.52HH277 pKa = 6.67DD278 pKa = 3.65MLMRR282 pKa = 11.84GSKK285 pKa = 9.49IGYY288 pKa = 7.92KK289 pKa = 9.98RR290 pKa = 11.84PVYY293 pKa = 9.67MYY295 pKa = 9.99GGSGSKK301 pKa = 10.4LVGLIEE307 pKa = 4.04
Molecular weight: 35.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1662 |
66 |
635 |
237.4 |
26.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.498 ± 0.941 | 1.083 ± 0.477 |
6.438 ± 1.098 | 4.091 ± 0.407 |
5.475 ± 1.003 | 6.318 ± 0.761 |
1.504 ± 0.377 | 6.619 ± 2.016 |
4.693 ± 0.718 | 9.146 ± 1.175 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.407 ± 0.376 | 4.994 ± 0.604 |
4.272 ± 0.987 | 4.031 ± 1.227 |
5.596 ± 0.868 | 10.168 ± 1.272 |
5.355 ± 0.721 | 5.054 ± 0.741 |
1.143 ± 0.329 | 5.114 ± 0.966 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
